Mutational analysis of highly conserved aspartate residues essential to the catalytic core of the piggyBac transposase
- PMID: 18694512
- PMCID: PMC2533014
- DOI: 10.1186/1471-2199-9-73
Mutational analysis of highly conserved aspartate residues essential to the catalytic core of the piggyBac transposase
Abstract
Background: The piggyBac mobile element is quickly gaining popularity as a tool for the transgenesis of many eukaryotic organisms. By studying the transposase which catalyzes the movement of piggyBac, we may be able to modify this vector system to make it a more effective transgenesis tool. In a previous publication, Sarkar A, Sim C, Hong YS, Hogan JR, Fraser MJ, Robertson HM, and Collins FH have proposed the presence of the widespread 'DDE/DDD' motif for piggyBac at amino acid positions D268, D346, and D447.
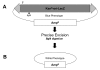
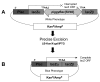
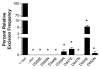
Results: This study utilizes directed mutagenesis and plasmid-based mobility assays to assess the importance of these residues as the catalytic core of the piggyBac transposase. We have functionally analyzed individual point-mutations with respect to charge and physical size in all three proposed residues of the 'DDD' motif as well as another nearby, highly conserved aspartate at D450. All of our mutations had a significant effect on excision frequency in S2 cell cultures. We have also aligned the piggyBac transposase to other close family members, both functional and non-functional, in an attempt to identify the most highly conserved regions and position a number of interesting features.
Conclusion: We found all the designated DDD aspartates reside in clusters of amino acids that conserved among piggyBac family transposase members. Our results indicate that all four aspartates are necessary, to one degree or another, for excision to occur in a cellular environment, but D450 seems to have a tolerance for a glutamate substitution. All mutants tested significantly decreased excision frequency in cell cultures when compared with the wild-type transposase.
Figures










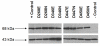
References
-
- Engels WR. P Elements in Drosophila. In: Saedler H, Gierl A, editor. Transposable Elements. Berlin , Springer; 1996. pp. 103–123.
-
- Craig NL. Mobile DNA II. Washington, D.C. , ASM Press; 2002. p. xviii, 1204 p., [32] p. of plates.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

