New time-scale criteria for model simplification of bio-reaction systems
- PMID: 18694523
- PMCID: PMC2553091
- DOI: 10.1186/1471-2105-9-338
New time-scale criteria for model simplification of bio-reaction systems
Abstract
Background: Quasi-steady state approximation (QSSA) based on time-scale analysis is known to be an effective method for simplifying metabolic reaction system, but the conventional analysis becomes time-consuming and tedious when the system is large. Although there are automatic methods, they are based on eigenvalue calculations of the Jacobian matrix and on linear transformations, which have a high computation cost. A more efficient estimation approach is necessary for complex systems.
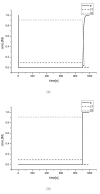
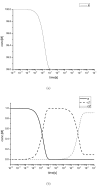
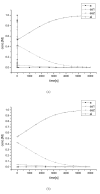
Results: This work derived new time-scale factor by focusing on the problem structure. By mathematically reasoning the balancing behavior of fast species, new time-scale criteria were derived with a simple expression that uses the Jacobian matrix directly. The algorithm requires no linear transformation or decomposition of the Jacobian matrix, which has been an essential part for previous automatic time-scaling methods. Furthermore, the proposed scale factor is estimated locally. Therefore, an iterative procedure was also developed to find the possible multiple boundary layers and to derive an appropriate reduced model.
Conclusion: By successive calculation of the newly derived time-scale criteria, it was possible to detect multiple boundary layers of full ordinary differential equation (ODE) models. Besides, the iterative procedure could derive the appropriate reduced differential algebraic equation (DAE) model with consistent initial values, which was tested with simple examples and a practical example.
Figures

Similar articles
-
Sensitivity analysis of dynamic biological systems with time-delays.BMC Bioinformatics. 2010 Oct 15;11 Suppl 7(Suppl 7):S12. doi: 10.1186/1471-2105-11-S7-S12. BMC Bioinformatics. 2010. PMID: 21106119 Free PMC article.
-
Reduction of multiscale stochastic biochemical reaction networks using exact moment derivation.PLoS Comput Biol. 2017 Jun 5;13(6):e1005571. doi: 10.1371/journal.pcbi.1005571. eCollection 2017 Jun. PLoS Comput Biol. 2017. PMID: 28582397 Free PMC article.
-
Scalable Parameter Estimation for Genome-Scale Biochemical Reaction Networks.PLoS Comput Biol. 2017 Jan 23;13(1):e1005331. doi: 10.1371/journal.pcbi.1005331. eCollection 2017 Jan. PLoS Comput Biol. 2017. PMID: 28114351 Free PMC article.
-
A structured approach for the engineering of biochemical network models, illustrated for signalling pathways.Brief Bioinform. 2008 Sep;9(5):404-21. doi: 10.1093/bib/bbn026. Epub 2008 Jun 23. Brief Bioinform. 2008. PMID: 18573813 Review.
-
Optimization in computational systems biology.BMC Syst Biol. 2008 May 28;2:47. doi: 10.1186/1752-0509-2-47. BMC Syst Biol. 2008. PMID: 18507829 Free PMC article. Review.
Cited by
-
Model reduction in mathematical pharmacology : Integration, reduction and linking of PBPK and systems biology models.J Pharmacokinet Pharmacodyn. 2018 Aug;45(4):537-555. doi: 10.1007/s10928-018-9584-y. Epub 2018 Mar 26. J Pharmacokinet Pharmacodyn. 2018. PMID: 29582349 Free PMC article.
-
A combined model reduction algorithm for controlled biochemical systems.BMC Syst Biol. 2017 Feb 13;11(1):17. doi: 10.1186/s12918-017-0397-1. BMC Syst Biol. 2017. PMID: 28193218 Free PMC article.
-
BioUML: an integrated environment for systems biology and collaborative analysis of biomedical data.Nucleic Acids Res. 2019 Jul 2;47(W1):W225-W233. doi: 10.1093/nar/gkz440. Nucleic Acids Res. 2019. PMID: 31131402 Free PMC article.
-
Model composition through model reduction: a combined model of CD95 and NF-κB signaling pathways.BMC Syst Biol. 2013 Feb 15;7:13. doi: 10.1186/1752-0509-7-13. BMC Syst Biol. 2013. PMID: 23409788 Free PMC article.
-
Methods of Model Reduction for Large-Scale Biological Systems: A Survey of Current Methods and Trends.Bull Math Biol. 2017 Jul;79(7):1449-1486. doi: 10.1007/s11538-017-0277-2. Epub 2017 Jun 27. Bull Math Biol. 2017. PMID: 28656491 Free PMC article.
References
-
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IκB-NF-κB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. - PubMed
-
- Dahlquist G, Edsberg L, Sköllermo G, Söderlind G. Are the numerical methods and software satisfactory for chemical kinetics? Lect Notes Math. 1982;968:149–164.
-
- Schauer M, Heinrich R. Quasi-steady-state approximation in the mathematical modelling of biochemical reaction networks. Math Biosci. 1983;65:155–170.
-
- Schuster R, Schuster S, Holzhutter HG. Simplification of complex kinetic models used for the quantitative analysis of nuclear magnetic resonance or Radioactive Tracer Studies. J Chem Soc Faraday Trans. 1992;88:2837–2844.
-
- Schuster R, Holzhutter HG. Rapid-equilibrium approximation applied to mathematical models of tracer dynamics in biochemical systems. Mathl Comput Modelling. 1994;19:241–253.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

