Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells
- PMID: 18694959
- PMCID: PMC2577430
- DOI: 10.1128/MCB.00923-08
Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells
Abstract
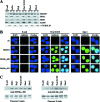
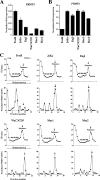
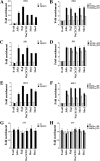
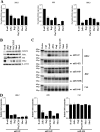
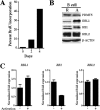
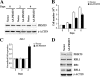
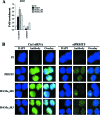
The proper epigenetic modification of chromatin by protein arginine methyltransferases (PRMTs) is crucial for normal cell growth and health. The human SWI/SNF-associated PRMT5 is involved in the transcriptional repression of target genes by directly methylating H3R8 and H4R3. To further understand the impact of PRMT5-mediated histone methylation on cancer, we analyzed its expression in normal and transformed human B lymphocytes. Our findings reveal that PRMT5 protein levels are enhanced in various human lymphoid cancer cells, including transformed chronic lymphocytic leukemia (B-CLL) cell lines. PRMT5 overexpression is caused by the altered expression of the PRMT5-specific microRNAs 19a, 25, 32, 92, 92b, and 96 and results in the increased global symmetric methylation of H3R8 and H4R3. An evaluation of both epigenetic marks at PRMT5 target genes such as RB1 (p105), RBL1 (p107), and RBL2 (p130) showed that promoters H3R8 and H4R3 are hypermethylated, which in turn triggers pocket protein transcriptional repression. Furthermore, reducing PRMT5 expression in WaC3CD5 B-CLL cells abolishes H3R8 and H4R3 hypermethylation, restores RBL2 expression, and inhibits cancer cell proliferation. These results indicate that PRMT5 overexpression epigenetically alters the transcription of key tumor suppressor genes and suggest a causal role of the elevated symmetric methylation of H3R8 and H4R3 at the RBL2 promoter in transformed B-lymphocyte pathology.
Figures


References
-
- Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. - PubMed
-
- Bedford, M. T., and S. Richard. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell 18263-272. - PubMed
-
- Brehm, A., and T. Kouzarides. 1999. Retinoblastoma protein meets chromatin. Trends Biochem. Sci. 24142-145. - PubMed
-
- Chapman, E. J., and J. C. Carrington. 2007. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8884-896. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
