Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation
- PMID: 18695247
- PMCID: PMC2575290
- DOI: 10.1073/pnas.0804621105
Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation
Abstract
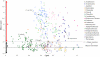
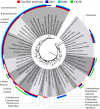
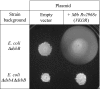
Protein disulfide bond formation contributes to the folding and activity of many exported proteins in bacteria. However, information about disulfide bond formation is limited to only a few bacterial species. We used a multifaceted bioinformatic approach to assess the capacity for disulfide bond formation across this biologically diverse group of organisms. We combined data from a cysteine counting method, in which a significant bias for even numbers of cysteine in proteins is taken as an indicator of disulfide bond formation, with data on the presence of homologs of known disulfide bond formation enzymes. These combined data enabled us to make predictions about disulfide bond formation in the cell envelope across bacterial species. Our bioinformatic and experimental results suggest that many bacteria may not generally oxidatively fold proteins, and implicate the bacterial homolog of the enzyme vitamin K epoxide reductase, a protein required for blood clotting in humans, as part of a disulfide bond formation pathway present in several major bacterial phyla.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Metabolism of long-chain fatty acids affects disulfide bond formation in Escherichia coli and activates envelope stress response pathways as a combat strategy.PLoS Genet. 2020 Oct 20;16(10):e1009081. doi: 10.1371/journal.pgen.1009081. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33079953 Free PMC article.
-
Identification of the Thioredoxin Partner of Vitamin K Epoxide Reductase in Mycobacterial Disulfide Bond Formation.J Bacteriol. 2018 Jul 25;200(16):e00137-18. doi: 10.1128/JB.00137-18. Print 2018 Aug 15. J Bacteriol. 2018. PMID: 29784887 Free PMC article.
-
Disulfide bond formation in prokaryotes.Nat Microbiol. 2018 Mar;3(3):270-280. doi: 10.1038/s41564-017-0106-2. Epub 2018 Feb 20. Nat Microbiol. 2018. PMID: 29463925 Review.
-
Exploring synonymous codon usage preferences of disulfide-bonded and non-disulfide bonded cysteines in the E. coli genome.J Theor Biol. 2006 Jul 21;241(2):390-401. doi: 10.1016/j.jtbi.2005.12.004. Epub 2006 Jan 19. J Theor Biol. 2006. PMID: 16427089
-
Mechanisms of oxidative protein folding in the bacterial cell envelope.Antioxid Redox Signal. 2010 Oct;13(8):1231-46. doi: 10.1089/ars.2010.3187. Antioxid Redox Signal. 2010. PMID: 20367276 Free PMC article. Review.
Cited by
-
Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.Protein Cell. 2013 Aug;4(8):628-40. doi: 10.1007/s13238-013-3033-x. Epub 2013 Jul 5. Protein Cell. 2013. PMID: 23828196 Free PMC article.
-
Virtual Screening of Peptide and Peptidomimetic Fragments Targeted to Inhibit Bacterial Dithiol Oxidase DsbA.PLoS One. 2015 Jul 30;10(7):e0133805. doi: 10.1371/journal.pone.0133805. eCollection 2015. PLoS One. 2015. PMID: 26225423 Free PMC article.
-
Metabolism of long-chain fatty acids affects disulfide bond formation in Escherichia coli and activates envelope stress response pathways as a combat strategy.PLoS Genet. 2020 Oct 20;16(10):e1009081. doi: 10.1371/journal.pgen.1009081. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33079953 Free PMC article.
-
Phylogenetically informed logic relationships improve detection of biological network organization.BMC Bioinformatics. 2011 Dec 15;12:476. doi: 10.1186/1471-2105-12-476. BMC Bioinformatics. 2011. PMID: 22172058 Free PMC article.
-
Dissecting the machinery that introduces disulfide bonds in Pseudomonas aeruginosa.mBio. 2013 Dec 10;4(6):e00912-13. doi: 10.1128/mBio.00912-13. mBio. 2013. PMID: 24327342 Free PMC article.
References
-
- Nakamoto H, Bardwell JC. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim Biophys Acta. 2004;1694:111–119. - PubMed
-
- Sone M, Kishigami S, Yoshihisa T, Ito K. Roles of disulfide bonds in bacterial alkaline phosphatase. J Biol Chem. 1997;272:6174–6178. - PubMed
-
- Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35–44. - PubMed
-
- Pollock MR, Richmond MH. Low cyst (e) ine content of bacterial extracellular proteins: Its possible physiological significance. Nature. 1962;194:446–449. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

