The ocular albinism type 1 protein, an intracellular G protein-coupled receptor, regulates melanosome transport in pigment cells
- PMID: 18697795
- PMCID: PMC2572695
- DOI: 10.1093/hmg/ddn241
The ocular albinism type 1 protein, an intracellular G protein-coupled receptor, regulates melanosome transport in pigment cells
Erratum in
-
The ocular albinism type 1 protein, an intracellular G protein-coupled receptor, regulates melanosome transport in pigment cells.Hum Mol Genet. 2017 Aug 1;26(15):3028-3029. doi: 10.1093/hmg/ddx131. Hum Mol Genet. 2017. PMID: 28854565 Free PMC article. No abstract available.
Abstract
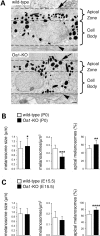
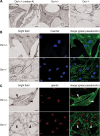
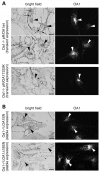
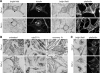
The protein product of the ocular albinism type 1 gene, named OA1, is a pigment cell-specific G protein-coupled receptor exclusively localized to intracellular organelles, namely lysosomes and melanosomes. Loss of OA1 function leads to the formation of macromelanosomes, suggesting that this receptor is implicated in organelle biogenesis, however the mechanism involved in the pathogenesis of the disease remains obscure. We report here the identification of an unexpected abnormality in melanosome distribution both in retinal pigment epithelium (RPE) and skin melanocytes of Oa1-knock-out (KO) mice, consisting in a displacement of the organelles from the central cytoplasm towards the cell periphery. Despite their depletion from the microtubule (MT)-enriched perinuclear region, Oa1-KO melanosomes were able to aggregate at the centrosome upon disruption of the actin cytoskeleton or expression of a dominant-negative construct of myosin Va. Consistently, quantification of organelle transport in living cells revealed that Oa1-KO melanosomes displayed a severe reduction in MT-based motility; however, this defect was rescued to normal following inhibition of actin-dependent capture at the cell periphery. Together, these data point to a defective regulation of organelle transport in the absence of OA1 and imply that the cytoskeleton might represent a downstream effector of this receptor. Furthermore, our results enlighten a novel function for OA1 in pigment cells and suggest that ocular albinism type 1 might result from a different pathogenetic mechanism than previously thought, based on an organelle-autonomous signalling pathway implicated in the regulation of both membrane traffic and transport.
Figures




References
-
- Quevedo W.C., Fitzpatrick T.B., Szabo G., Jimbow K. Biology of melanocytes. In: Fitzpatrick T.B., Eisen A.Z., Wolff K., Freedberg I.M., Austen K.F., editors. Dermatology in General Medicine. Vol. 1. New York: McGraw-Hill; 1987. pp. 224–251.
-
- Orlow S.J. Melanosomes are specialized members of the lysosomal lineage of organelles. J. Invest. Dermatol. 1995;105:3–7. - PubMed
-
- Dell'Angelica E.C., Mullins C., Caplan S., Bonifacino J.S. Lysosome-related organelles. FASEB J. 2000;14:1265–1278. - PubMed
-
- Raposo G., Marks M.S. The dark side of lysosome-related organelles: specialization of the endocytic pathway for melanosome biogenesis. Traffic. 2002;3:237–248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

