Mutations in JARID1C are associated with X-linked mental retardation, short stature and hyperreflexia
- PMID: 18697827
- PMCID: PMC3711528
- DOI: 10.1136/jmg.2008.058990
Mutations in JARID1C are associated with X-linked mental retardation, short stature and hyperreflexia
Abstract
Background: Mutations in the JARID1C (Jumonji AT-rich interactive domain 1C) gene were recently associated with X-linked mental retardation (XLMR). Mutations in this gene are reported to be one of the relatively more common causes of XLMR with a frequency of approximately 3% in males with proven or probable XLMR. The JARID1C protein functions as a histone 3 lysine 4 (H3K4) demethylase and is involved in the demethylation of H3K4me3 and H3K4me2.
Methods: Mutation analysis of the JARID1C gene was conducted in the following cohorts: probands from 23 XLMR families linked to Xp11.2, 92 males with mental retardation and short stature, and 172 probands from small XLMR families with no linkage information.
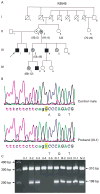
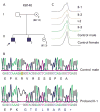
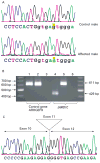
Results: Four novel mutations consisting of two missense mutations, p.A77T and p.V504M, and two frame shift mutations, p.E468fsX2 and p.R1481fsX9, were identified in males with mental retardation. Two of the mutations, p.V504M and p.E468fsX2, are located in the JmjC domain of the JARID1C gene where no previous mutations have been reported. Additional studies showed that the missense mutation, p.V504M, was a de novo event on the grandpaternal X chromosome of the family. Clinical findings of the nine affected males from the four different families included mental retardation (100%), short stature (55%), hyperreflexia (78%), seizures (33%) and aggressive behaviour (44%). The degree of mental retardation consisted of mild (25%), moderate (12%) and severe (63%).
Conclusion: Based on the clinical observations, male patients with mental retardation, short stature and hyperreflexia should be considered candidates for mutations in the JARID1C gene.
Conflict of interest statement
Figures


References
-
- Schalock RL, Luckasson RA, Shogren KA, Borthwick-Duffy S, Bradley V, Buntinx WH, Coulter DL, Craig EM, Gomez SC, Lachapelle Y, Reeve A, Snell ME, Spreat S, Tassé MJ, Thompson JR, Verdugo MA, Wehmeyer ML, Yeager MH. The renaming of mental retardation: understanding the change to the term intellectual disability. Intellect Dev Disabil. 2007;45:116–24. - PubMed
-
- Larson SA, Lakin KC, Anderson L, Kwak Lee N, Anderson D. Prevalence of mental retardation and developmental disabilities: estimates from the 1994/1995 national health interview survey disability supplements. Am J Ment Retard. 2001;106:231–52. - PubMed
-
- Brosco JP, Mattingly M, Sanders LM. Impact of specific medical interventions on reducing the prevalence of mental retardation. Arch Pediatr Adolesc Med. 2006;160:302–9. - PubMed
-
- Stevenson RE. Advances in X-linked mental retardation. Curr Opin Pediatr. 2005;17:720–4. - PubMed
-
- Stevenson RE, Schwartz CE, Schroer RJ. X-linked mental retardation. New York: Oxford University Press; 2000.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
