Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs
- PMID: 18697922
- PMCID: PMC2525944
- DOI: 10.1261/rna.1062908
Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs
Abstract
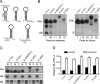
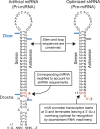
RNA interference (RNAi) is a cellular process regulating gene expression and participating in innate defense in many organisms. RNAi has also been utilized as a tool to query gene function and is being developed as a therapeutic strategy for several diseases. Synthetic small interfering (siRNAs) or expressed stem-loop RNAs (short-hairpin RNAs [shRNAs] or artificial microRNAs [miRNAs]) have been delivered to cultured cells and organisms to inhibit expression of a variety of genes. A persistent question in the field, however, is which RNAi expression system is most suitable for distinct applications. To date, shRNA- and artificial miRNA-based strategies have been compared with conflicting results. In prior comparisons, sequences required for efficient RNAi processing and loading of the intended antisense strand into the RNAi-induced silencing complex (RISC) were not considered. We therefore revisited the shRNA-miRNA comparison question. Initially, we developed an improved artificial miRNA vector and confirmed the optimal shRNA configuration by altering structural features of these RNAi substrates. Subsequently, we engineered and compared shRNA- and miRNA-based RNAi expression vectors that would be processed to yield similar siRNAs that exhibit comparable strand biasing. Our results demonstrate that when comparison variables are minimized, the shRNAs tested were more potent than the artificial miRNAs in mediating gene silencing independent of target sequence and experimental setting (in vitro and in vivo). In addition, we show that shRNAs are expressed at considerably higher levels relative to artificial miRNAs, thus providing mechanistic insight to explain their increased potency.
Figures

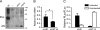
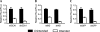
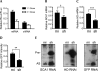
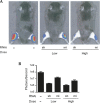
References
-
- Birmingham A., Anderson E.M., Reynolds A., Ilsley-Tyree D., Leake D., Fedorov Y., Baskerville S., Maksimova E., Robinson K., Karpilow J., et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. - PubMed
-
- Bridge A.J., Pebernard S., Ducraux A., Nicoulaz A.L., Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
