The leaf ionome as a multivariable system to detect a plant's physiological status
- PMID: 18697928
- PMCID: PMC2515221
- DOI: 10.1073/pnas.0804175105
The leaf ionome as a multivariable system to detect a plant's physiological status
Abstract
The contention that quantitative profiles of biomolecules contain information about the physiological state of the organism has motivated a variety of high-throughput molecular profiling experiments. However, unbiased discovery and validation of biomolecular signatures from these experiments remains a challenge. Here we show that the Arabidopsis thaliana (Arabidopsis) leaf ionome, or elemental composition, contains such signatures, and we establish statistical models that connect these multivariable signatures to defined physiological responses, such as iron (Fe) and phosphorus (P) homeostasis. Iron is essential for plant growth and development, but potentially toxic at elevated levels. Because of this, shoot Fe concentrations are tightly regulated and show little variation over a range of Fe concentrations in the environment, making them a poor probe of a plant's Fe status. By evaluating the shoot ionome in plants grown under different Fe nutritional conditions, we have established a multivariable ionomic signature for the Fe response status of Arabidopsis. This signature has been validated against known Fe-response proteins and allows the high-throughput detection of the Fe status of plants with a false negative/positive rate of 18%/16%. A "metascreen" of previously collected ionomic data from 880 Arabidopsis mutants and natural accessions for this Fe response signature successfully identified the known Fe mutants frd1 and frd3. A similar approach has also been taken to identify and use a shoot ionomic signature associated with P homeostasis. This study establishes that multivariable ionomic signatures of physiological states associated with mineral nutrient homeostasis do exist in Arabidopsis and are in principle robust enough to detect specific physiological responses to environmental or genetic perturbations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
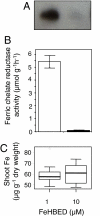
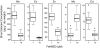


Comment in
-
Validation of multivariate model of leaf ionome is fundamentally confounded.Proc Natl Acad Sci U S A. 2009 Jan 13;106(2):E6; author reply E7. doi: 10.1073/pnas.0809853106. Epub 2008 Dec 31. Proc Natl Acad Sci U S A. 2009. PMID: 19118190 Free PMC article. No abstract available.
References
-
- Gerszten RE, Wang TJ. The search for new cardiovascular biomarkers. Nature. 2008;451:949–952. - PubMed
-
- Ward M. Biomarkers for Alzheimer's disease. Exp Rev Mol Diagn. 2007;7:635–646. - PubMed
-
- Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

