In vitro analysis of integrated global high-resolution DNA methylation profiling with genomic imbalance and gene expression in osteosarcoma
- PMID: 18698372
- PMCID: PMC2515339
- DOI: 10.1371/journal.pone.0002834
In vitro analysis of integrated global high-resolution DNA methylation profiling with genomic imbalance and gene expression in osteosarcoma
Abstract
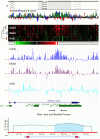
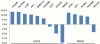
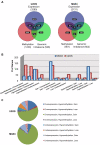
Genetic and epigenetic changes contribute to deregulation of gene expression and development of human cancer. Changes in DNA methylation are key epigenetic factors regulating gene expression and genomic stability. Recent progress in microarray technologies resulted in developments of high resolution platforms for profiling of genetic, epigenetic and gene expression changes. OS is a pediatric bone tumor with characteristically high level of numerical and structural chromosomal changes. Furthermore, little is known about DNA methylation changes in OS. Our objective was to develop an integrative approach for analysis of high-resolution epigenomic, genomic, and gene expression profiles in order to identify functional epi/genomic differences between OS cell lines and normal human osteoblasts. A combination of Affymetrix Promoter Tilling Arrays for DNA methylation, Agilent array-CGH platform for genomic imbalance and Affymetrix Gene 1.0 platform for gene expression analysis was used. As a result, an integrative high-resolution approach for interrogation of genome-wide tumour-specific changes in DNA methylation was developed. This approach was used to provide the first genomic DNA methylation maps, and to identify and validate genes with aberrant DNA methylation in OS cell lines. This first integrative analysis of global cancer-related changes in DNA methylation, genomic imbalance, and gene expression has provided comprehensive evidence of the cumulative roles of epigenetic and genetic mechanisms in deregulation of gene expression networks.
Conflict of interest statement
Figures





Similar articles
-
Integrative analysis reveals relationships of genetic and epigenetic alterations in osteosarcoma.PLoS One. 2012;7(11):e48262. doi: 10.1371/journal.pone.0048262. Epub 2012 Nov 7. PLoS One. 2012. PMID: 23144859 Free PMC article.
-
Epigenetic mapping and functional analysis in a breast cancer metastasis model using whole-genome promoter tiling microarrays.Breast Cancer Res. 2008;10(4):R62. doi: 10.1186/bcr2121. Epub 2008 Jul 18. Breast Cancer Res. 2008. PMID: 18638373 Free PMC article.
-
Analysis of miRNA-gene expression-genomic profiles reveals complex mechanisms of microRNA deregulation in osteosarcoma.Cancer Genet. 2011 Mar;204(3):138-46. doi: 10.1016/j.cancergen.2010.12.012. Cancer Genet. 2011. PMID: 21504713
-
Comparison of genome-wide analysis techniques to DNA methylation analysis in human cancer.J Cell Physiol. 2018 May;233(5):3968-3981. doi: 10.1002/jcp.26176. Epub 2017 Oct 4. J Cell Physiol. 2018. PMID: 28888056 Review.
-
Genome-wide DNA methylation profiling.Wiley Interdiscip Rev Syst Biol Med. 2010 Mar-Apr;2(2):210-223. doi: 10.1002/wsbm.35. Wiley Interdiscip Rev Syst Biol Med. 2010. PMID: 20836023 Review.
Cited by
-
Wilms' tumor gene 1 silencing inhibits proliferation of human osteosarcoma MG-63 cell line by cell cycle arrest and apoptosis activation.Oncotarget. 2017 Feb 21;8(8):13917-13931. doi: 10.18632/oncotarget.14715. Oncotarget. 2017. PMID: 28107196 Free PMC article.
-
Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers.Reprod Toxicol. 2012 Dec;34(4):694-707. doi: 10.1016/j.reprotox.2012.09.005. Epub 2012 Oct 2. Reprod Toxicol. 2012. PMID: 23041264 Free PMC article.
-
Batch effects and pathway analysis: two potential perils in cancer studies involving DNA methylation array analysis.Cancer Epidemiol Biomarkers Prev. 2013 Jun;22(6):1052-60. doi: 10.1158/1055-9965.EPI-13-0114. Epub 2013 Apr 29. Cancer Epidemiol Biomarkers Prev. 2013. PMID: 23629520 Free PMC article.
-
Integrated DNA Copy Number and Expression Profiling Identifies IGF1R as a Prognostic Biomarker in Pediatric Osteosarcoma.Int J Mol Sci. 2022 Jul 21;23(14):8036. doi: 10.3390/ijms23148036. Int J Mol Sci. 2022. PMID: 35887382 Free PMC article.
-
New cytogenetically visible copy number variant in region 8q21.2.Mol Cytogenet. 2011 Jan 5;4(1):1. doi: 10.1186/1755-8166-4-1. Mol Cytogenet. 2011. PMID: 21208402 Free PMC article.
References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. - PubMed
-
- Momparler RL, Bovenzi V. DNA methylation and cancer. J Cell Physiol. 2000;183:145–154. - PubMed
-
- Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

