Identifying determinants of cullin binding specificity among the three functionally different Drosophila melanogaster Roc proteins via domain swapping
- PMID: 18698375
- PMCID: PMC2500221
- DOI: 10.1371/journal.pone.0002918
Identifying determinants of cullin binding specificity among the three functionally different Drosophila melanogaster Roc proteins via domain swapping
Abstract
Background: Cullin-dependent E3 ubiquitin ligases (CDL) are key regulators of protein destruction that participate in a wide range of cell biological processes. The Roc subunit of CDL contains an evolutionarily conserved RING domain that binds ubiquitin charged E2 and is essential for ubiquitylation. Drosophila melanogaster contains three highly related Roc proteins: Roc1a and Roc2, which are conserved in vertebrates, and Roc1b, which is specific to Drosophila. Our previous genetic data analyzing Roc1a and Roc1b mutants suggested that Roc proteins are functionally distinct, but the molecular basis for this distinction is not known.
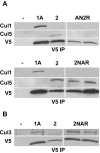
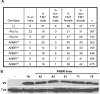
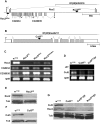
Methodology/principal findings: Using co-immunoprecipitation studies we show that Drosophila Roc proteins bind specific Cullins: Roc1a binds Cul1-4, Roc1b binds Cul3, and Roc2 binds Cul5. Through domain swapping experiments, we demonstrate that Cullin binding specificity is strongly influenced by the Roc NH(2)-terminal domain, which forms an inter-molecular beta sheet with the Cullin. Substitution of the Roc1a RING domain with that of Roc1b results in a protein with similar Cullin binding properties to Roc1a that is active as an E3 ligase but cannot complement Roc1a mutant lethality, indicating that the identity of the RING domain can be an important determinant of CDL function. In contrast, the converse chimeric protein with a substitution of the Roc1b RING domain with that of Roc1a can rescue the male sterility of Roc1b mutants, but only when expressed from the endogenous Roc1b promoter. We also identified mutations of Roc2 and Cul5 and show that they cause no overt developmental phenotype, consistent with our finding that Roc2 and Cul5 proteins are exclusive binding partners, which others have observed in human cells as well.
Conclusions: The Drosophila Roc proteins are highly similar, but have diverged during evolution to bind a distinct set of Cullins and to utilize RING domains that have overlapping, but not identical, function in vivo.
Conflict of interest statement
Figures




Similar articles
-
Targeted disruption of Drosophila Roc1b reveals functional differences in the Roc subunit of Cullin-dependent E3 ubiquitin ligases.Mol Biol Cell. 2004 Nov;15(11):4892-903. doi: 10.1091/mbc.e04-03-0180. Epub 2004 Aug 25. Mol Biol Cell. 2004. PMID: 15331761 Free PMC article.
-
A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila.PLoS Biol. 2007 Oct;5(10):e251. doi: 10.1371/journal.pbio.0050251. PLoS Biol. 2007. PMID: 17880263 Free PMC article.
-
Regulation of Drosophila vasa in vivo through paralogous cullin-RING E3 ligase specificity receptors.Mol Cell Biol. 2010 Apr;30(7):1769-82. doi: 10.1128/MCB.01100-09. Epub 2010 Feb 1. Mol Cell Biol. 2010. PMID: 20123973 Free PMC article.
-
Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family.EMBO J. 2004 Apr 21;23(8):1681-7. doi: 10.1038/sj.emboj.7600186. Epub 2004 Apr 8. EMBO J. 2004. PMID: 15071497 Free PMC article. Review.
-
CRL4s: the CUL4-RING E3 ubiquitin ligases.Trends Biochem Sci. 2009 Nov;34(11):562-70. doi: 10.1016/j.tibs.2009.07.002. Epub 2009 Oct 7. Trends Biochem Sci. 2009. PMID: 19818632 Free PMC article. Review.
Cited by
-
Peripheral Neuropathy and Decreased Locomotion of a RAB40B Mutation in Human and Model Animals.Exp Neurobiol. 2023 Dec 31;32(6):410-422. doi: 10.5607/en23027. Exp Neurobiol. 2023. PMID: 38196136 Free PMC article.
-
Functional characterization of SAG/RBX2/ROC2/RNF7, an antioxidant protein and an E3 ubiquitin ligase.Protein Cell. 2013 Feb;4(2):103-16. doi: 10.1007/s13238-012-2105-7. Epub 2012 Nov 8. Protein Cell. 2013. PMID: 23136067 Free PMC article. Review.
-
Cullin-5 and cullin-2 play a role in the development of neuromuscular junction and the female germ line of Drosophila.J Genet. 2011 Aug;90(2):239-49. doi: 10.1007/s12041-011-0062-1. J Genet. 2011. PMID: 21869472
-
Small RING Finger Proteins RBX1 and RBX2 of SCF E3 Ubiquitin Ligases: The Role in Cancer and as Cancer Targets.Genes Cancer. 2010 Jul;1(7):700-7. doi: 10.1177/1947601910382776. Genes Cancer. 2010. PMID: 21103004 Free PMC article.
-
The ubiquitin-conjugating enzyme UBE2E3 and its import receptor importin-11 regulate the localization and activity of the antioxidant transcription factor NRF2.Mol Biol Cell. 2015 Jan 15;26(2):327-38. doi: 10.1091/mbc.E14-06-1057. Epub 2014 Nov 5. Mol Biol Cell. 2015. PMID: 25378586 Free PMC article.
References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. - PubMed
-
- Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. - PubMed
-
- Pelzer C, Kassner I, Matentzoglu K, Singh RK, Wollscheid HP, et al. UBE1L2, a novel E1 enzyme specific for ubiquitin. J Biol Chem. 2007;282:23010–23014. - PubMed
-
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

