Convergent energy and stress signaling
- PMID: 18701338
- PMCID: PMC3075853
- DOI: 10.1016/j.tplants.2008.06.006
Convergent energy and stress signaling
Abstract
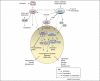
Plants are constantly confronted by multiple types of stress. Despite their distinct origin and mode of perception, nutrient deprivation and most stresses have an impact on the overall energy status of the plant, leading to convergent downstream responses that include largely overlapping transcriptional patterns. The emerging view is that this transcriptome reprogramming in energy and stress signaling is partly regulated by the evolutionarily conserved energy sensor protein kinases, SNF1 (sucrose non-fermenting 1) in yeast, AMPK (AMP-activated protein kinase) in mammals and SnRK1 (SNF1-related kinase 1) in plants. Upon sensing the energy deficit associated with stress, nutrient deprivation and darkness, SnRK1 triggers extensive transcriptional changes that contribute to restoring homeostasis, promoting cell survival and elaborating longer-term responses for adaptation, growth and development.
Figures


References
-
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. - PubMed
-
- Baena-Gonzalez E, et al. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. - PubMed
-
- Smith AM, Stitt M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007;30:1126–1149. - PubMed
-
- Kilian J, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

