Combined genetic and modeling approaches reveal that epidermal cell area and number in leaves are controlled by leaf and plant developmental processes in Arabidopsis
- PMID: 18701672
- PMCID: PMC2556812
- DOI: 10.1104/pp.108.124271
Combined genetic and modeling approaches reveal that epidermal cell area and number in leaves are controlled by leaf and plant developmental processes in Arabidopsis
Abstract
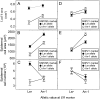
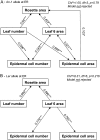
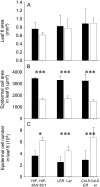
Both leaf production and leaf expansion are tightly linked to cell expansion and cell division, but the functional relationships between all these variables are not clearly established. To get insight into these relationships, a quantitative genetic analysis was performed in 118 recombinant inbred lines derived from a cross between the Landsberg erecta and Antwerp accessions and was combined with a structural equation modeling approach. Main effects and epistatic interactions at the quantitative trait locus (QTL) level were detected for rosette area, rosette leaf number, leaf 6 area, epidermal cell area and number. A QTL at ERECTA marker (ER) controlled cell expansion and cell division, in interaction with two other QTLs at SNP295 and SNP21 markers. Moreover, both the screening for marker association involved in the variation of the relationships between leaf growth variables and the test of alternative functional models by structural equation modeling revealed that the allelic value at ER controlled epidermal cell area and epidermal cell number in a leaf. These effects are driven both by a whole plant mechanism associated with leaf production and by a single leaf mechanism associated with leaf expansion. The complex effects of the QTL at ER were validated in selected heterogeneous inbred families. The ERECTA gene, which is mutated in the Landsberg erecta parental line, was found to be a putative candidate responsible for these mapped effects by phenotyping mutants of this gene at the cellular level. Together, these results give insight into the complex determination of leaf epidermal cell number and area.
Figures
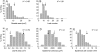


References
-
- Aguirrezabal L, Bouchier-Combaud S, Radziejwoski A, Dauzat M, Cookson SJ, Granier C (2006) Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ 29 2216–2227 - PubMed
-
- Ashby E (1948) Studies in the morphogenesis of leaves. 2. The area, cell size and cell number of leaves of Ipomoea in relation to their position on the shoot. New Phytol 47 177–195
-
- Chase K, Adler FR, Lark KG (1997) Epistat: a computer program for identifying and testing interactions between pairs of quantitative trait loci. Theor Appl Genet 94 724–730
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

