Calnexin is involved in apoptosis induced by endoplasmic reticulum stress in the fission yeast
- PMID: 18701708
- PMCID: PMC2555920
- DOI: 10.1091/mbc.e08-02-0188
Calnexin is involved in apoptosis induced by endoplasmic reticulum stress in the fission yeast
Abstract
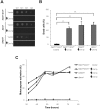
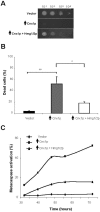
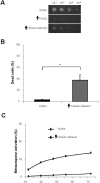
Stress conditions affecting the functions of the endoplasmic reticulum (ER) cause the accumulation of unfolded proteins. ER stress is counteracted by the unfolded-protein response (UPR). However, under prolonged stress the UPR initiates a proapoptotic response. Mounting evidence indicate that the ER chaperone calnexin is involved in apoptosis caused by ER stress. Here, we report that overexpression of calnexin in Schizosaccharomyces pombe induces cell death with apoptosis markers. Cell death was partially dependent on the Ire1p ER-stress transducer. Apoptotic death caused by calnexin overexpression required its transmembrane domain (TM), and involved sequences on either side of the ER membrane. Apoptotic death caused by tunicamycin was dramatically reduced in a strain expressing endogenous levels of calnexin lacking its TM and cytosolic tail. This demonstrates the involvement of calnexin in apoptosis triggered by ER stress. A genetic screen identified the S. pombe homologue of the human antiapoptotic protein HMGB1 as a suppressor of apoptotic death due to calnexin overexpression. Remarkably, overexpression of human calnexin in S. pombe also provoked apoptotic death. Our results argue for the conservation of the role of calnexin in apoptosis triggered by ER stress, and validate S. pombe as a model to elucidate the mechanisms of calnexin-mediated cell death.
Figures
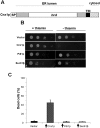
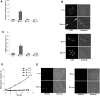

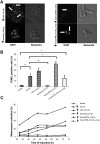
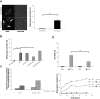
Similar articles
-
Calnexin regulates apoptosis induced by inositol starvation in fission yeast.PLoS One. 2009 Jul 16;4(7):e6244. doi: 10.1371/journal.pone.0006244. PLoS One. 2009. PMID: 19606215 Free PMC article.
-
Cell viability and secretion of active proteins in Schizosaccharomyces pombe do not require the chaperone function of calnexin.Biochem J. 2004 Jun 1;380(Pt 2):441-8. doi: 10.1042/BJ20031546. Biochem J. 2004. PMID: 14984369 Free PMC article.
-
A non-chromosomal factor allows viability of Schizosaccharomyces pombe lacking the essential chaperone calnexin.J Cell Sci. 2004 Feb 29;117(Pt 6):907-18. doi: 10.1242/jcs.00943. J Cell Sci. 2004. PMID: 14963023
-
The cellular response to protein misfolding in the endoplasmic reticulum.Gene Expr. 1999;7(4-6):293-300. Gene Expr. 1999. PMID: 10440230 Free PMC article. Review.
-
Calnexin phosphorylation: linking cytoplasmic signalling to endoplasmic reticulum lumenal functions.Semin Cell Dev Biol. 2010 Jul;21(5):486-90. doi: 10.1016/j.semcdb.2009.12.005. Epub 2010 Jan 6. Semin Cell Dev Biol. 2010. PMID: 20005969 Review.
Cited by
-
Coordination of the unfolded protein response during hepatic steatosis identifies CHOP as a specific regulator of hepatocyte ballooning.Cell Stress Chaperones. 2020 Nov;25(6):969-978. doi: 10.1007/s12192-020-01132-x. Epub 2020 Jun 23. Cell Stress Chaperones. 2020. PMID: 32577989 Free PMC article.
-
Yeast as a tool to decipher the molecular mechanisms underlying the functions of Bcl-2 family.Explor Target Antitumor Ther. 2022;3(2):128-148. doi: 10.37349/etat.2022.00076. Epub 2022 Apr 2. Explor Target Antitumor Ther. 2022. PMID: 36046841 Free PMC article. Review.
-
Endoplasmic reticulum involvement in yeast cell death.Front Oncol. 2012 Aug 2;2:87. doi: 10.3389/fonc.2012.00087. eCollection 2012. Front Oncol. 2012. PMID: 22876361 Free PMC article.
-
Upregulation of the Rab27a-dependent trafficking and secretory mechanisms improves lysosomal transport, alleviates endoplasmic reticulum stress, and reduces lysosome overload in cystinosis.Mol Cell Biol. 2013 Aug;33(15):2950-62. doi: 10.1128/MCB.00417-13. Epub 2013 May 28. Mol Cell Biol. 2013. PMID: 23716592 Free PMC article.
-
MCP-1/MCPIP-1 Signaling Modulates the Effects of IL-1β in Renal Cell Carcinoma through ER Stress-Mediated Apoptosis.Int J Mol Sci. 2019 Dec 3;20(23):6101. doi: 10.3390/ijms20236101. Int J Mol Sci. 2019. PMID: 31816951 Free PMC article.
References
-
- Ahner A., Brodsky J. L. Checkpoints in ER-associated degradation: excuse me, which way to the proteasome? Trends Cell Biol. 2004;14:474–478. - PubMed
-
- Almeida B., Silva A., Mesquita A., Sampaio-Marques B., Rodrigues F., Ludovico P. Drug-induced apoptosis in yeast. Biochim. Biophys. Acta. 2008;1783:1436–1448. - PubMed
-
- Arunachalam B., Cresswell P. Molecular requirements for the interaction of class II major histocompatibility complex molecules and invariant chain with calnexin. J. Biol. Chem. 1995;270:2784–2790. - PubMed
-
- Barbet N., Muriel W. J., Carr A. M. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

