Double-strand breaks associated with repetitive DNA can reshape the genome
- PMID: 18701715
- PMCID: PMC2515620
- DOI: 10.1073/pnas.0804529105
Double-strand breaks associated with repetitive DNA can reshape the genome
Abstract
Ionizing radiation is an established source of chromosome aberrations (CAs). Although double-strand breaks (DSBs) are implicated in radiation-induced and other CAs, the underlying mechanisms are poorly understood. Here, we show that, although the vast majority of randomly induced DSBs in G(2) diploid yeast cells are repaired efficiently through homologous recombination (HR) between sister chromatids or homologous chromosomes, approximately 2% of all DSBs give rise to CAs. Complete molecular analysis of the genome revealed that nearly all of the CAs resulted from HR between nonallelic repetitive elements, primarily Ty retrotransposons. Nonhomologous end-joining (NHEJ) accounted for few, if any, of the CAs. We conclude that only those DSBs that fall at the 3-5% of the genome composed of repetitive DNA elements are efficient at generating rearrangements with dispersed small repeats across the genome, whereas DSBs in unique sequences are confined to recombinational repair between the large regions of homology contained in sister chromatids or homologous chromosomes. Because repeat-associated DSBs can efficiently lead to CAs and reshape the genome, they could be a rich source of evolutionary change.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
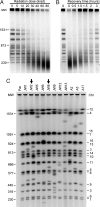


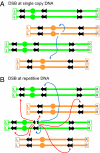
Comment in
-
From the shards of a shattered genome, diversity.Proc Natl Acad Sci U S A. 2008 Aug 19;105(33):11593-4. doi: 10.1073/pnas.0805812105. Epub 2008 Aug 13. Proc Natl Acad Sci U S A. 2008. PMID: 18701721 Free PMC article. No abstract available.
References
-
- Muller HJ. Artificial transmutation of the gene. Science. 1927;66:84–87. - PubMed
-
- McClintock B. Cytological observations of deficiencies involving known genes, translocations and inversions in Zea mays. MO Agric Exp Station Res Bull. 1931;163:1–30.
-
- Kupiec M. Damage-induced recombination in the yeast Saccharomyces cerevisiae. Mutat Res. 2000;451:91–105. - PubMed
-
- Fasullo M, Dave P, Rothstein R. DNA-damaging agents stimulate the formation of directed reciprocal translocations in Saccharomyces cerevisiae. Mutat Res. 1994;314:121–133. - PubMed
-
- Myung K, Kolodner RD. Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2003;2:243–258. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

