SUMO modification of PCNA is controlled by DNA
- PMID: 18701921
- PMCID: PMC2518684
- DOI: 10.1038/emboj.2008.162
SUMO modification of PCNA is controlled by DNA
Abstract
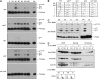
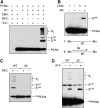
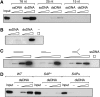
Post-translational modification by the ubiquitin-like protein SUMO is often regulated by cellular signals that restrict the modification to appropriate situations. Nevertheless, many SUMO-specific ligases do not exhibit much target specificity, and--compared with the diversity of sumoylation substrates--their number is limited. This raises the question of how SUMO conjugation is controlled in vivo. We report here an unexpected mechanism by which sumoylation of the replication clamp protein, PCNA, from budding yeast is effectively coupled to S phase. We find that loading of PCNA onto DNA is a prerequisite for sumoylation in vivo and greatly stimulates modification in vitro. To our surprise, however, DNA binding by the ligase Siz1, responsible for PCNA sumoylation, is not strictly required. Instead, the stimulatory effect of DNA on conjugation is mainly attributable to DNA binding of PCNA itself. These findings imply a change in the properties of PCNA upon loading that enhances its capacity to be sumoylated.
Figures



References
-
- Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtsuka E, Tsurimoto T (1995) Structure–function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. J Biol Chem 270: 22527–22534 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

