Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: ex vivo effects and in vivo "traces" on the murine IAPE and human HERV-K elements
- PMID: 18702815
- PMCID: PMC2531183
- DOI: 10.1186/1742-4690-5-75
Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: ex vivo effects and in vivo "traces" on the murine IAPE and human HERV-K elements
Abstract
Background: APOBEC3 cytosine deaminases have been demonstrated to restrict infectivity of a series of retroviruses, with different efficiencies depending on the retrovirus. In addition, APOBEC3 proteins can severely restrict the intracellular transposition of a series of retroelements with a strictly intracellular life cycle, including the murine IAP and MusD LTR-retrotransposons.
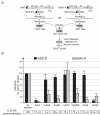
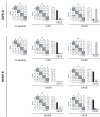
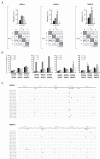
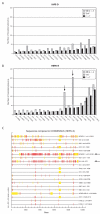
Results: Here we show that the IAPE element, which is the infectious progenitor of the strictly intracellular IAP elements, and the infectious human endogenous retrovirus HERV-K are restricted by both murine and human APOBEC3 proteins in an ex vivo assay for infectivity, with evidence in most cases of strand-specific G-to-A editing of the proviruses, with the expected signatures. In silico analysis of the naturally occurring genomic copies of the corresponding endogenous elements performed on the mouse and human genomes discloses "traces" of APOBEC3-editing, with the specific signature of the murine APOBEC3 and human APOBEC3G enzymes, respectively, and to a variable extent depending on the family member.
Conclusion: These results indicate that the IAPE and HERV-K elements, which can only replicate via an extracellular infection cycle, have been restricted at the time of their entry, amplification and integration into their target host genomes by definite APOBEC3 proteins, most probably acting in evolution to limit the mutagenic effect of these endogenized extracellular parasites.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

