TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing
- PMID: 18703504
- PMCID: PMC2661999
- DOI: 10.1074/jbc.M805376200
TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing
Abstract
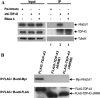
TDP-43 is a highly conserved, 43-kDa RNA-binding protein implicated to play a role in transcription repression, nuclear organization, and alternative splicing. More recently, this factor has been identified as the major disease protein of several neurodegenerative diseases, including frontotemporal lobar degeneration with ubiquitin-positive inclusions and amyotrophic lateral sclerosis. For the splicing activity, the factor has been shown to be mainly an exon-skipping promoter. In this study using the survival of motor neuron (SMN) minigenes as the reporters in transfection assay, we show for the first time that TDP-43 could also act as an exon-inclusion factor. Furthermore, both RNA-recognition motif domains are required for its ability to enhance the SMN2 exon 7 inclusion. Combined protein-immunoprecipitation and RNA-immunoprecipitation experiments also suggested that this exon inclusion activity might be mediated by multimeric complex(es) consisting of this protein interacting with other splicing factors, including Htra2-beta1. Our data further evidence TDP-43 as a multifunctional RNA-binding protein for a diverse set of cellular activities.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

