Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome
- PMID: 18703679
- PMCID: PMC2532931
- DOI: 10.1101/gad.1682308
Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome
Abstract
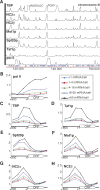
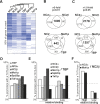
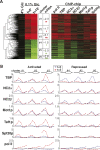
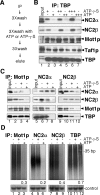
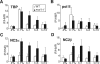
Promoter recognition by TATA-binding protein (TBP) is an essential step in the initiation of RNA polymerase II (pol II) mediated transcription. Genetic and biochemical studies in yeast have shown that Mot1p and NC2 play important roles in inhibiting TBP activity. To understand how TBP activity is regulated in a genome-wide manner, we profiled the binding of TBP, NC2, Mot1p, TFIID, SAGA, and pol II across the yeast genome using chromatin immunoprecipitation (ChIP)-chip for cells in exponential growth and during reprogramming of transcription. We find that TBP, NC2, and Mot1p colocalize at transcriptionally active pol II core promoters. Relative binding of NC2alpha and Mot1p is higher at TATA promoters, whereas NC2beta has a preference for TATA-less promoters. In line with the ChIP-chip data, we isolated a stable TBP-NC2-Mot1p-DNA complex from chromatin extracts. ATP hydrolysis releases NC2 and DNA from the Mot1p-TBP complex. In vivo experiments indicate that promoter dissociation of TBP and NC2 is highly dynamic, which is dependent on Mot1p function. Based on these results, we propose that NC2 and Mot1p cooperate to dynamically restrict TBP activity on transcribed promoters.
Figures
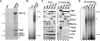
References
-
- Auble D.T., Hansen K.E., Mueller C.G., Lane W.S., Thorner J., Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes & Dev. 1994;8:1920–1934. - PubMed
-
- Basehoar A.D., Zanton S.J., Pugh B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. - PubMed
-
- Blake W.J., Balazsi G., Kohanski M.A., Isaacs F.J., Murphy K.F., Kuang Y., Cantor C.R., Walt D.R., Collins J.J. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell. 2006;24:853–865. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
