Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase
- PMID: 18704088
- PMCID: PMC2597408
- DOI: 10.1038/nature07162
Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase
Abstract
Non-ribosomal peptide synthetases (NRPS) and polyketide synthases (PKS) produce numerous secondary metabolites with various therapeutic/antibiotic properties. Like fatty acid synthases (FAS), these enzymes are organized in modular assembly lines in which each module, made of conserved domains, incorporates a given monomer unit into the growing chain. Knowledge about domain or module interactions may enable reengineering of this assembly line enzymatic organization and open avenues for the design of new bioactive compounds with improved therapeutic properties. So far, little structural information has been available on how the domains interact and communicate. This may be because of inherent interdomain mobility hindering crystallization, or because crystallized molecules may not represent the active domain orientations. In solution, the large size and internal dynamics of multidomain fragments (>35 kilodaltons) make structure determination by nuclear magnetic resonance a challenge and require advanced technologies. Here we present the solution structure of the apo-thiolation-thioesterase (T-TE) di-domain fragment of the Escherichia coli enterobactin synthetase EntF NRPS subunit. In the holoenzyme, the T domain carries the growing chain tethered to a 4'-phosphopantetheine whereas the TE domain catalyses hydrolysis and cyclization of the iron chelator enterobactin. The T-TE di-domain forms a compact but dynamic structure with a well-defined domain interface; the two active sites are at a suitable distance for substrate transfer from T to TE. We observe extensive interdomain and intradomain motions for well-defined regions and show that these are modulated by interactions with proteins that participate in the biosynthesis. The T-TE interaction described here provides a model for NRPS, PKS and FAS function in general as T-TE-like di-domains typically catalyse the last step in numerous assembly-line chain-termination machineries.
Figures
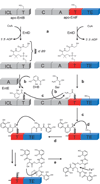


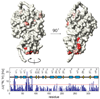
Comment in
-
Biochemistry: Fit for an enzyme.Nature. 2008 Aug 14;454(7206):832-3. doi: 10.1038/454832a. Nature. 2008. PMID: 18704072 No abstract available.
Similar articles
-
The EntF and EntE adenylation domains of Escherichia coli enterobactin synthetase: sequestration and selectivity in acyl-AMP transfers to thiolation domain cosubstrates.Proc Natl Acad Sci U S A. 2000 Mar 14;97(6):2509-14. doi: 10.1073/pnas.040572897. Proc Natl Acad Sci U S A. 2000. PMID: 10688898 Free PMC article.
-
Aminoacyl-SNACs as small-molecule substrates for the condensation domains of nonribosomal peptide synthetases.Chem Biol. 2000 Oct;7(10):765-72. doi: 10.1016/s1074-5521(00)00022-3. Chem Biol. 2000. PMID: 11033080
-
Dissection of the EntF condensation domain boundary and active site residues in nonribosomal peptide synthesis.Biochemistry. 2003 Feb 11;42(5):1334-44. doi: 10.1021/bi026867m. Biochemistry. 2003. PMID: 12564937
-
Protein-protein interactions in multienzyme megasynthetases.Chembiochem. 2008 Apr 14;9(6):826-48. doi: 10.1002/cbic.200700751. Chembiochem. 2008. PMID: 18357594 Review.
-
The many faces and important roles of protein-protein interactions during non-ribosomal peptide synthesis.Nat Prod Rep. 2018 Nov 14;35(11):1120-1139. doi: 10.1039/c8np00038g. Nat Prod Rep. 2018. PMID: 30207358 Review.
Cited by
-
Structural insight into the necessary conformational changes of modular nonribosomal peptide synthetases.Curr Opin Chem Biol. 2016 Dec;35:89-96. doi: 10.1016/j.cbpa.2016.09.005. Epub 2016 Sep 25. Curr Opin Chem Biol. 2016. PMID: 27676239 Free PMC article. Review.
-
Structural analysis of protein-protein interactions in type I polyketide synthases.Crit Rev Biochem Mol Biol. 2013 Mar-Apr;48(2):98-122. doi: 10.3109/10409238.2012.745476. Epub 2012 Dec 19. Crit Rev Biochem Mol Biol. 2013. PMID: 23249187 Free PMC article. Review.
-
Portability of the thiolation domain in recombinant pyoverdine non-ribosomal peptide synthetases.BMC Microbiol. 2015 Aug 13;15:162. doi: 10.1186/s12866-015-0496-3. BMC Microbiol. 2015. PMID: 26268580 Free PMC article.
-
Elucidation of novel turnagainolides and their biosynthetic gene cluster in Bacillus subtilis.Appl Environ Microbiol. 2025 May 21;91(5):e0257424. doi: 10.1128/aem.02574-24. Epub 2025 Apr 25. Appl Environ Microbiol. 2025. PMID: 40277365 Free PMC article.
-
New insights into the formation of fungal aromatic polyketides.Nat Rev Microbiol. 2010 Dec;8(12):879-89. doi: 10.1038/nrmicro2465. Nat Rev Microbiol. 2010. PMID: 21079635 Free PMC article. Review.
References
-
- Walsh CT. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303:1805–1810. - PubMed
-
- Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen LO. Structural and functional insights into a peptide bond-forming bidomain from a nonribosomal peptide synthetase. Structure. 2007;15:781–792. - PubMed
-
- Gehring AM, Bradley KA, Walsh CT. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36:8495–8503. - PubMed
-
- Gehring AM, Mori I, Walsh CT. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry. 1998;37:2648–2659. - PubMed
-
- Wagner G. The importance of being floppy. Nature Struct. Biol. 1995;2:255–257. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

