Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers
- PMID: 18704501
- PMCID: PMC2525844
- DOI: 10.1007/s00439-008-0535-3
Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers
Abstract
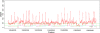
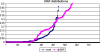
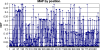
Recently, genome-wide association studies have identified loci across a segment of chromosome 8q24 (128,100,000-128,700,000) associated with the risk of breast, colon and prostate cancers. At least three regions of 8q24 have been independently associated with prostate cancer risk; the most centromeric of which appears to be population specific. Haplotypes in two contiguous but independent loci, marked by rs6983267 and rs1447295, have been identified in the Cancer Genetic Markers of Susceptibility project ( http://cgems.cancer.gov ), which genotyped more than 5,000 prostate cancer cases and 5,000 controls of European origin. The rs6983267 locus is also strongly associated with colorectal cancer. To ascertain a comprehensive catalog of common single-nucleotide polymorphisms (SNPs) across the two regions, we conducted a resequence analysis of 136 kb (chr8: 128,473,000-128,609,802) using the Roche/454 next-generation sequencing technology in 39 prostate cancer cases and 40 controls of European origin. We have characterized a comprehensive catalog of common (MAF > 1%) SNPs within this region, including 442 novel SNPs and have determined the pattern of linkage disequilibrium across the region. Our study has generated a detailed map of genetic variation across the region, which should be useful for choosing SNPs for fine mapping of association signals in 8q24 and investigations of the functional consequences of select common variants.
Figures



References
-
- Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. - DOI - PubMed
-
- Berndt SI, Potter JD, Hazra A, Yeager M, Thomas G, Makar KW, Welch R, Cross AJ, Huang WY, Schoen RE, Giovannucci E, Chan AT, Chanock SJ, Peters U, Hunter DJ, Hayes RB (2008) Pooled analysis of genetic variation at chromosome 8q24 and colorectal neoplasia risk. Hum Mol Genet. doi:10.1093/hmg/ddn166 - PMC - PubMed
-
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr, Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

