Lettuce mosaic virus: from pathogen diversity to host interactors
- PMID: 18705846
- PMCID: PMC6640324
- DOI: 10.1111/j.1364-3703.2007.00451.x
Lettuce mosaic virus: from pathogen diversity to host interactors
Abstract
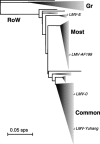
Taxonomy: Lettuce mosaic virus (LMV) belongs to the genus Potyvirus (type species Potato virus Y) in the family Potyviridae.
Physical properties: The virion is filamentous, flexuous with a length of 750 nm and a width of 15 nm. The particles are made of a genomic RNA of 10 080 nucleotides, covalently linked to a viral-encoded protein (the VPg) at the 5' end and with a 3' poly A tail, and encapsidated in a single type of capsid protein. The molecular weight of the capsid protein subunit has been estimated electrophoretically to be 34 kDa and estimated from the amino acid sequence to be 31 kDa.
Genome organization: The genome is expressed as a polyprotein of 3255 amino-acid residues, processed by three virus-specific proteinases into ten mature proteins.
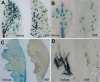
Hosts: LMV has a worldwide distribution and a relatively broad host range among several families. Weeds and ornamentals can act as local reservoirs for lettuce crops. In particular, many species within the family Asteraceae are susceptible to LMV, including cultivated and ornamental species such as common (Lactuca sativa), prickly (L. serriola) or wild (L. virosa) lettuce, endive/escarole (Cichorium endiva), safflower (Carthamus tinctorius), starthistle (Centaurea solstitialis), Cape daisy (Osteospermum spp.) and gazania (Gazania rigens). In addition, several species within the families Brassicaceae, Cucurbitaceae, Fabaceae, Solanaceae and Chenopodiaceae are natural or experimental hosts of LMV. Genetic control of resistance to LMV: The only resistance genes currently used to protect lettuce crops worldwide are the recessive genes mo1(1) and mo1(2) corresponding to mutant alleles of the gene encoding the translation initiation factor eIF4E in lettuce. It is believed that at least one intact copy of eIF4E must be present to ensure virus accumulation.
Transmission: LMV is transmitted in a non-persistent manner by a high number of aphid species. Myzus persicae and Macrosiphum euphorbiae are particularly active in disseminating this virus in the fields. LMV is also seedborne in lettuce. The effectiveness of LMV transmission depends on the cultivar and the age of the seed carrier at the inoculation time.
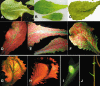
Symptoms: The characteristic symptoms on susceptible lettuce cultivars are dwarfism, mosaic, distortion and yellowing of the leaves with sometimes a much reduced heart of lettuce (failure to form heads). The differences in virus strains, cultivars and the physiological stage of the host at the moment of the attack cause different symptom severity: from a very slight discoloration of the veins to severe necrosis leading to the death of the plant.
Figures

Similar articles
-
The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus Lettuce mosaic virus.Plant Physiol. 2003 Jul;132(3):1272-82. doi: 10.1104/pp.102.017855. Plant Physiol. 2003. PMID: 12857809 Free PMC article.
-
Comparison of biological and molecular characterization of Iranian lettuce mosaic virus isolates.Commun Agric Appl Biol Sci. 2006;71(3 Pt B):1289-94. Commun Agric Appl Biol Sci. 2006. PMID: 17390892
-
The C terminus of lettuce mosaic potyvirus cylindrical inclusion helicase interacts with the viral VPg and with lettuce translation eukaryotic initiation factor 4E.J Gen Virol. 2012 Jan;93(Pt 1):184-193. doi: 10.1099/vir.0.035881-0. Epub 2011 Sep 14. J Gen Virol. 2012. PMID: 21918009
-
Potato virus Y: a major crop pathogen that has provided major insights into the evolution of viral pathogenicity.Mol Plant Pathol. 2013 Jun;14(5):439-52. doi: 10.1111/mpp.12024. Epub 2013 Mar 11. Mol Plant Pathol. 2013. PMID: 23480826 Free PMC article. Review.
-
Pepino mosaic virus: a successful pathogen that rapidly evolved from emerging to endemic in tomato crops.Mol Plant Pathol. 2010 Mar;11(2):179-89. doi: 10.1111/j.1364-3703.2009.00600.x. Mol Plant Pathol. 2010. PMID: 20447268 Free PMC article. Review.
Cited by
-
Quantitative and qualitative involvement of P3N-PIPO in overcoming recessive resistance against Clover yellow vein virus in pea carrying the cyv1 gene.J Virol. 2013 Jul;87(13):7326-37. doi: 10.1128/JVI.00065-13. Epub 2013 Apr 24. J Virol. 2013. PMID: 23616656 Free PMC article.
-
A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection.Plant Physiol. 2010 Jan;152(1):255-66. doi: 10.1104/pp.109.147983. Epub 2009 Oct 30. Plant Physiol. 2010. PMID: 19880609 Free PMC article.
-
Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E-mediated resistance against Lettuce mosaic potyvirus.Mol Plant Pathol. 2009 Jan;10(1):109-13. doi: 10.1111/j.1364-3703.2008.00513.x. Mol Plant Pathol. 2009. PMID: 19161357 Free PMC article.
-
Genome-wide association study of the seed transmission rate of soybean mosaic virus and associated traits using two diverse population panels.Theor Appl Genet. 2019 Dec;132(12):3413-3424. doi: 10.1007/s00122-019-03434-w. Epub 2019 Oct 19. Theor Appl Genet. 2019. PMID: 31630210
-
Differential mRNA Accumulation upon Early Arabidopsis thaliana Infection with ORMV and TMV-Cg Is Associated with Distinct Endogenous Small RNAs Level.PLoS One. 2015 Aug 3;10(8):e0134719. doi: 10.1371/journal.pone.0134719. eCollection 2015. PLoS One. 2015. PMID: 26237414 Free PMC article.
References
-
- Adams, M.J. , Antoniw, J.F. and Fauquet, C.M. (2005) Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 150, 459–479. - PubMed
-
- Arroyo, R. , Soto, M.J. , Martinez‐Zapater, J.M. and Ponz, F. (1996) Impaired cell‐to‐cell movement of Potato Virus Y in pepper plants carrying the ya (pvr2 1) resistance gene. Mol. Plant–Microbe Interact. 9, 314–318.
-
- Ayme, V. , Petit‐Pierre, J. , Souche, S. , Palloix, A. and Moury, B. (2007) Molecular dissection of the potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 88, 1594–1601. - PubMed
-
- Ayme, V. , Souche, S. , Caranta, C. , Jacquemond, M. , Chadoeuf, J. , Palloix, A. and Moury, B. (2006) Different mutations in the genome‐linked protein VPg of potato virus Y confer virulence on the pvr2(3) resistance in pepper. Mol. Plant–Microbe Interact. 19, 557–563. - PubMed
-
- Bannerot, H. , Boulidard, L. , Marrou, J. and Duteil, M. (1969) Etude de la tolérance au virus de la mosaïque de la laitue chez la variété Gallega de Invierno. Ann. Phytopathol. 1, 219–226.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

