Transcriptome analysis of the wheat-Puccinia striiformis f. sp. tritici interaction
- PMID: 18705849
- PMCID: PMC6640478
- DOI: 10.1111/j.1364-3703.2007.00453.x
Transcriptome analysis of the wheat-Puccinia striiformis f. sp. tritici interaction
Abstract
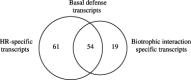
Stripe rust [caused by Puccinia striiformis Westend. f. sp. tritici Eriks. (Pst)] is a destructive disease of wheat (Triticum aestivum L.) worldwide. Genetic resistance is the preferred method for control and the Yr5 gene, originally identified in Triticum spelta var. album, represents a major resistance (R) gene that confers all-stage resistance to all currently known races of Pst in the United States. To identify transcripts associated with the Yr5-mediated incompatible interaction and the yr5-compatible interaction, the Wheat GeneChip was used to profile the changes occurring in wheat isolines that differed for the presence of the Yr5 gene after inoculation with Pst. This time-course study (6, 12, 24 and 48 h post-inoculation) identified 115 transcripts that were induced during the R-gene-mediated incompatible interaction, and 73 induced during the compatible interaction. Fifty-four transcripts were induced in both interactions and were considered as basal defence transcripts, whilst 61 transcripts were specific to the incompatible interaction [hypersensitive response (HR)-specific transcripts] and 19 were specific to the compatible interaction (biotrophic interaction-specific transcripts). The temporal pattern of transcript accumulation showed a peak at 24 h after infection that may reflect haustorial penetration by Pst at ~16 h. An additional 12 constitutive transcript differences were attributed to the presence of Yr5 after eliminating those considered as incomplete isogenicity. Annotation of the induced transcripts revealed that the presence of Yr5 resulted in a rapid and amplified resistance response involving signalling pathways and defence-related transcripts known to occur during R-gene-mediated responses, including protein kinase signalling and the production of reactive oxygen species leading to a hypersensitive response. Basal defence also involved substantial induction of many defence-related transcripts but the lack of R-gene signalling resulted in weaker response.
Figures
References
-
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 57, 289–300.
-
- Bindschedler, L. , Dewdney, J. , Blee, K. , Stone, J. , Asai, T. , Plotnikov, J. , Denoux, C. , Hayes, T. , Gerrish, C. , Davies, D.R. , Ausubel, F.M. and Bolwell, G.P. (2006) Peroxidase‐dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 47, 851–863. - PMC - PubMed
-
- Boddu, J. , Cho, S. , Kruger, W. and Muehlbauer, G. (2006) Transcriptome analysis of the barley‐Fusarium graminearum interaction. Mol. Plant–Microbe Interact. 19, 407–417. - PubMed
-
- Bolstad, B. , Irizarry, R. , Astrand, M. and Speed, T. (2003) A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics, 19, 185–193. - PubMed
-
- Bouche, N. , Yellin, A. , Snedden, W. and Fromm, H. (2005) Plant‐specific calmodulin‐binding proteins. Annu. Rev. Plant Biol. 56, 435–466. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

