Histological and molecular analysis of Rdg2a barley resistance to leaf stripe
- PMID: 18705861
- PMCID: PMC6640343
- DOI: 10.1111/j.1364-3703.2008.00479.x
Histological and molecular analysis of Rdg2a barley resistance to leaf stripe
Abstract
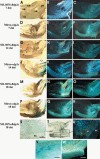
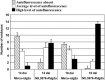
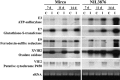
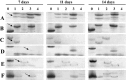
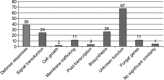
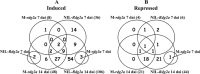
Barley (Hordeum vulgare L.) leaf stripe is caused by the seed-borne fungus Pyrenophora graminea. We investigated microscopically and molecularly the reaction of barley embryos to leaf stripe inoculation. In the resistant genotype NIL3876-Rdg2a, fungal growth ceased at the scutellar node of the embryo, while in the susceptible near-isogenic line (NIL) Mirco-rdg2a fungal growth continued past the scutellar node and into the embryo. Pathogen-challenged embryos of resistant and susceptible NILs showed different levels of UV autofluorescence and toluidine blue staining, indicating differential accumulation of phenolic compounds. Suppression subtractive hybridization and cDNA amplified fragment-length polymorphism (AFLP) analyses of embryos identified P. graminea-induced and P. graminea-repressed barley genes. In addition, cDNA-AFLP analysis identified six pathogenicity-associated fungal genes expressed during barley infection but at low to undetectable levels during growth on artificial media. Microarrays representing the entire set of differentially expressed cDNA-AFLP fragments and 100 barley homologues of previously described defence-related genes were used to study gene expression changes at 7 and 14 days after inoculation in the resistant and susceptible NILs. A total of 171 significantly modulated barley genes were identified and assigned to four groups based on timing and genotype dependence of expression. Analysis of the changes in gene expression during the barley resistance response to leaf stripe suggests that the Rdg2a-mediated response includes cell-wall reinforcement, signal transduction, generation of reactive oxygen species, cell protection, jasmonate signalling and expression of plant effector genes. The identification of genes showing leaf stripe inoculation or resistance-dependent expression sets the stage for further dissection of the resistance response of barley embryo cells to leaf stripe.
Figures


References
-
- Aragona, M. and Porta‐Puglia, A. (1999) Identification of resistance to leaf stripe using a Pyrenophora graminea transformant expressing β‐glucuronidase. Eur. J. Plant Pathol. 105, 831–834.
-
- Arru, L. , Francia, E. and Pecchioni, N. (2003) Isolate‐specific QTLs of resistance to leaf stripe (Pyrenophora graminea) in the Steptoe × Morex spring barley cross. Theor. Appl. Genet. 106, 668–675. - PubMed
-
- Arru, L. , Nicks, R.E. , Lindhout, P. , Valè, G. , Francia, E. and Pecchioni, N. (2002) Genomic regions determining resistance to leaf stripe (Pyrenophora graminea) in barley. Genome, 45, 460–466. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

