Post-translational modification of host proteins in pathogen-triggered defence signalling in plants
- PMID: 18705867
- PMCID: PMC6640405
- DOI: 10.1111/j.1364-3703.2008.00468.x
Post-translational modification of host proteins in pathogen-triggered defence signalling in plants
Abstract
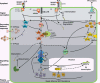
Microbial plant pathogens impose a continuous threat to global food production. Similar to animals, an innate immune system allows plants to recognize pathogens and swiftly activate defence. To activate a rapid response, receptor-mediated pathogen perception and subsequent downstream signalling depends on post-translational modification (PTM) of components essential for defence signalling. We discuss different types of PTMs that play a role in mounting plant immunity, which include phosphorylation, glycosylation, ubiquitination, sumoylation, nitrosylation, myristoylation, palmitoylation and glycosylphosphatidylinositol (GPI)-anchoring. PTMs are rapid, reversible, controlled and highly specific, and provide a tool to regulate protein stability, activity and localization. Here, we give an overview of PTMs that modify components essential for defence signalling at the site of signal perception, during secondary messenger production and during signalling in the cytoplasm. In addition, we discuss effectors from pathogens that suppress plant defence responses by interfering with host PTMs.
Figures
References
-
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. - PubMed
-
- Andersson, M.X. , Kourtchenko, O. , Dangl, J.L. , Mackey, D. and Ellerström, M. (2006) Phospholipase‐dependent signalling during the AvrRpm1‐ and AvrRpt2‐induced disease resistance responses in Arabidopsis thaliana . Plant J. 47, 947–959. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

