Decoding the quantitative nature of TGF-beta/Smad signaling
- PMID: 18706811
- PMCID: PMC2774497
- DOI: 10.1016/j.tcb.2008.06.006
Decoding the quantitative nature of TGF-beta/Smad signaling
Abstract
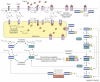
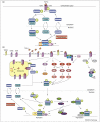
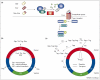
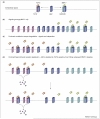
How transforming growth factor-beta (TGF-beta) signaling elicits diverse cell responses remains elusive, despite the major molecular components of the pathway being known. We contend that understanding TGF-beta biology requires mathematical models to decipher the quantitative nature of TGF-beta/Smad signaling and to account for its complexity. Here, we review mathematical models of TGF-beta superfamily signaling that predict how robustness is achieved in bone-morphogenetic-protein signaling in the Drosophila embryo, how changes in receptor-trafficking dynamics can be exploited by cancer cells and how the basic mechanisms of TGF-beta/Smad signaling conspire to promote Smad accumulation in the nucleus. These studies demonstrate the power of mathematical modeling for understanding TGF-beta biology.
Figures
Similar articles
-
Quantitative modeling and analysis of the transforming growth factor beta signaling pathway.Biophys J. 2009 Mar 4;96(5):1733-50. doi: 10.1016/j.bpj.2008.11.050. Biophys J. 2009. PMID: 19254534 Free PMC article.
-
Msk is required for nuclear import of TGF-{beta}/BMP-activated Smads.J Cell Biol. 2007 Sep 10;178(6):981-94. doi: 10.1083/jcb.200703106. Epub 2007 Sep 4. J Cell Biol. 2007. PMID: 17785517 Free PMC article.
-
A road map toward defining the role of Smad signaling in hematopoietic stem cells.Stem Cells. 2006 Apr;24(4):1128-36. doi: 10.1634/stemcells.2005-0263. Epub 2005 Dec 15. Stem Cells. 2006. PMID: 16357343
-
Dynamics of TGF-β/Smad signaling.FEBS Lett. 2012 Jul 4;586(14):1921-8. doi: 10.1016/j.febslet.2012.03.063. Epub 2012 Apr 9. FEBS Lett. 2012. PMID: 22710166 Free PMC article. Review.
-
TGF-β Family Signaling in Drosophila.Cold Spring Harb Perspect Biol. 2017 Sep 1;9(9):a022152. doi: 10.1101/cshperspect.a022152. Cold Spring Harb Perspect Biol. 2017. PMID: 28130362 Free PMC article. Review.
Cited by
-
Burst BMP triggered receptor kinase activity drives Smad1 mediated long-term target gene oscillation in C2C12 cells.PLoS One. 2013;8(4):e59442. doi: 10.1371/journal.pone.0059442. Epub 2013 Apr 1. PLoS One. 2013. PMID: 23560048 Free PMC article.
-
Chronic Stress Contributes to Osteosarcopenic Adiposity via Inflammation and Immune Modulation: The Case for More Precise Nutritional Investigation.Nutrients. 2020 Apr 2;12(4):989. doi: 10.3390/nu12040989. Nutrients. 2020. PMID: 32252359 Free PMC article. Review.
-
A high-throughput study on endothelial cell adhesion and growth mediated by adsorbed serum protein via signaling pathway PCR array.Regen Biomater. 2018 Feb;5(1):25-34. doi: 10.1093/rb/rbx030. Epub 2017 Dec 13. Regen Biomater. 2018. PMID: 29423265 Free PMC article.
-
Analyzing protein dynamics from fluorescence intensity traces using unsupervised deep learning network.Commun Biol. 2020 Nov 12;3(1):669. doi: 10.1038/s42003-020-01389-z. Commun Biol. 2020. PMID: 33184459 Free PMC article.
-
A crucial role for bone morphogenetic protein-Smad1 signalling in the DNA damage response.Nat Commun. 2012 May 15;3:836. doi: 10.1038/ncomms1832. Nat Commun. 2012. PMID: 22588298
References
-
- Inman GJ, et al. Nucleocytoplasmic shuttling of Smads 2, 3 and 4 permits sensing of TGF-β receptor activity. Mol. Cell. 2002;10:283–294. - PubMed
-
- Karlsson G, et al. Gene expression profiling demonstrates that TGF-β1 signals exclusively through receptor complexes involving Alk5 and identifies targets of TGF-β signaling. Physiol. Genomics. 2005;21:396–403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

