Decoding the quantitative nature of TGF-beta/Smad signaling
- PMID: 18706811
- PMCID: PMC2774497
- DOI: 10.1016/j.tcb.2008.06.006
Decoding the quantitative nature of TGF-beta/Smad signaling
Abstract
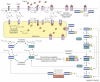
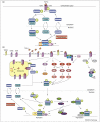
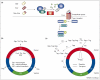
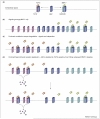
How transforming growth factor-beta (TGF-beta) signaling elicits diverse cell responses remains elusive, despite the major molecular components of the pathway being known. We contend that understanding TGF-beta biology requires mathematical models to decipher the quantitative nature of TGF-beta/Smad signaling and to account for its complexity. Here, we review mathematical models of TGF-beta superfamily signaling that predict how robustness is achieved in bone-morphogenetic-protein signaling in the Drosophila embryo, how changes in receptor-trafficking dynamics can be exploited by cancer cells and how the basic mechanisms of TGF-beta/Smad signaling conspire to promote Smad accumulation in the nucleus. These studies demonstrate the power of mathematical modeling for understanding TGF-beta biology.
Figures
References
-
- Inman GJ, et al. Nucleocytoplasmic shuttling of Smads 2, 3 and 4 permits sensing of TGF-β receptor activity. Mol. Cell. 2002;10:283–294. - PubMed
-
- Karlsson G, et al. Gene expression profiling demonstrates that TGF-β1 signals exclusively through receptor complexes involving Alk5 and identifies targets of TGF-β signaling. Physiol. Genomics. 2005;21:396–403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

