Quantifying protein interface footprinting by hydroxyl radical oxidation and molecular dynamics simulation: application to galectin-1
- PMID: 18707901
- PMCID: PMC2607067
- DOI: 10.1016/j.jasms.2008.07.013
Quantifying protein interface footprinting by hydroxyl radical oxidation and molecular dynamics simulation: application to galectin-1
Abstract
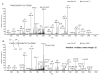
Biomolecular surface mapping methods offer an important alternative method for characterizing protein-protein and protein-ligand interactions in cases in which it is not possible to determine high-resolution three-dimensional (3D) structures of complexes. Hydroxyl radical footprinting offers a significant advance in footprint resolution compared with traditional chemical derivatization. Here we present results of footprinting performed with hydroxyl radicals generated on the nanosecond time scale by laser-induced photodissociation of hydrogen peroxide. We applied this emerging method to a carbohydrate-binding protein, galectin-1. Since galectin-1 occurs as a homodimer, footprinting was employed to characterize the interface of the monomeric subunits. Efficient analysis of the mass spectrometry data for the oxidized protein was achieved with the recently developed ByOnic (Palo Alto, CA) software that was altered to handle the large number of modifications arising from side-chain oxidation. Quantification of the level of oxidation has been achieved by employing spectral intensities for all of the observed oxidation states on a per-residue basis. The level of accuracy achievable from spectral intensities was determined by examination of mixtures of synthetic peptides related to those present after oxidation and tryptic digestion of galectin-1. A direct relationship between side-chain solvent accessibility and level of oxidation emerged, which enabled the prediction of the level of oxidation given the 3D structure of the protein. The precision of this relationship was enhanced through the use of average solvent accessibilities computed from 10 ns molecular dynamics simulations of the protein.
Figures
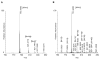
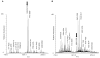

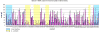
References
-
- King D, Bergmann C, Orlando R, Benen JA, Kester HC, Visser J. Use of Amide Exchange Mass Spectrometry to Study Conformational Changes Within the Endopolygalacturonase II-Homogalacturonan-Polygalacturonase Inhibiting Protein System. Biochemistry. 2002;41:10225–10233. - PubMed
-
- King D, Lumpkin M, Bergmann C, Orlando R. Studying Protein-Carbohydrate Interactions by Amide Hydrogen/Deuterium Exchange Mass Spectrometry. Rapid Commun Mass Spectrom. 2002;16:1569–1574. - PubMed
-
- Woods VL, Jr, Hamuro Y. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J Cell Biochem. 2001;Suppl 37:89–98. - PubMed
-
- Zheng X, Wintrode PL, Chance MR. Complementary structural mass spectrometry techniques reveal local dynamics in functionally important regions of a metastable serpin. Structure. 2008;16(1):38–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

