PDX-1 interaction and regulation of the Pancreatic Derived Factor (PANDER, FAM3B) promoter
- PMID: 18708173
- PMCID: PMC2574553
- DOI: 10.1016/j.bbagrm.2008.07.007
PDX-1 interaction and regulation of the Pancreatic Derived Factor (PANDER, FAM3B) promoter
Abstract
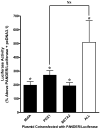
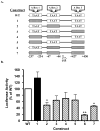
Pancreatic Derived Factor (PANDER) is a novel cytokine-like protein dominantly expressed within the endocrine pancreas. Our previous study demonstrated that the PANDER promoter was both tissue-specific and glucose-responsive. Surrounding the PANDER transcriptional start site are several putative A- and E-Box elements that may bind to the various pancreatic transcriptional factors of MafA, BETA2/NeuroD, and Pancreatic Duodenal Homeobox-1 (PDX-1). To characterize the transcriptional regulatory factors involved in PANDER gene expression, we performed co-transfection reporter gene analysis and demonstrated upregulation by all three transcription factors, with the greatest individual increase stemming from PDX-1. Potential binding of PDX-1 to A box (TAAT) regions of the PANDER promoter was demonstrated by chromatin immunoprecipitation (ChIP) and further corroborated by electrophoretic mobility shift assay (EMSA). Binding of PDX-1 to the A box regions was inhibited by mutagenized (TAGT) oligonucleotides. Site-directed mutagenesis of the three PDX-1 A box binding motifs revealed that A box sites 2 and 3 in combination were critical for maximal gene expression and deletion resulted in a 82% reduction in promoter activity. Furthermore, deletion of A box sites 2 and 3 completely diminished the glucose-responsiveness of the PANDER promoter. Our findings demonstrate that PANDER is a potential PDX-1 target gene and the A box sites within the promoter region are critical for basal and glucose-stimulated PANDER expression.
Figures



Similar articles
-
Hepatic nutrient and hormonal regulation of the PANcreatic-DERived factor (PANDER) promoter.Mol Cell Endocrinol. 2015 Sep 15;413:101-12. doi: 10.1016/j.mce.2015.05.040. Epub 2015 Jun 26. Mol Cell Endocrinol. 2015. PMID: 26123584
-
Tissue-specific and glucose-responsive expression of the pancreatic derived factor (PANDER) promoter.Biochim Biophys Acta. 2005 Sep 25;1730(3):215-25. doi: 10.1016/j.bbaexp.2005.07.003. Biochim Biophys Acta. 2005. PMID: 16102856
-
Regulation of pancreas duodenum homeobox-1 expression by early growth response-1.J Biol Chem. 2007 Mar 2;282(9):5973-83. doi: 10.1074/jbc.M607288200. Epub 2006 Dec 6. J Biol Chem. 2007. PMID: 17150967
-
Pancreatic duodenal homeobox factor-1 and diabetes mellitus type 2 (review).Int J Mol Med. 2008 Apr;21(4):399-404. Int J Mol Med. 2008. PMID: 18360684 Review.
-
Role of PDX-1 and MafA as a potential therapeutic target for diabetes.Diabetes Res Clin Pract. 2007 Sep;77 Suppl 1:S127-37. doi: 10.1016/j.diabres.2007.01.046. Epub 2007 Apr 20. Diabetes Res Clin Pract. 2007. PMID: 17449132 Review.
Cited by
-
Targeted disruption of pancreatic-derived factor (PANDER, FAM3B) impairs pancreatic beta-cell function.Diabetes. 2010 Sep;59(9):2209-18. doi: 10.2337/db09-1552. Epub 2010 Jun 21. Diabetes. 2010. PMID: 20566664 Free PMC article.
-
Circ_0134944 inhibits osteogenesis through miR-127-5p/PDX1/SPHK1 pathway.Regen Ther. 2021 Sep 29;18:391-400. doi: 10.1016/j.reth.2021.09.004. eCollection 2021 Dec. Regen Ther. 2021. PMID: 34722835 Free PMC article.
-
Quantitative proteomic profiling reveals hepatic lipogenesis and liver X receptor activation in the PANDER transgenic model.Mol Cell Endocrinol. 2016 Nov 15;436:41-9. doi: 10.1016/j.mce.2016.07.009. Epub 2016 Jul 7. Mol Cell Endocrinol. 2016. PMID: 27394190 Free PMC article.
-
New insights into the inter-organ crosstalk mediated by ChREBP.Front Endocrinol (Lausanne). 2023 Feb 27;14:1095440. doi: 10.3389/fendo.2023.1095440. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36923222 Free PMC article. Review.
-
Characterization of the expression, localization, and secretion of PANDER in alpha-cells.Mol Cell Endocrinol. 2010 Aug 30;325(1-2):36-45. doi: 10.1016/j.mce.2010.05.008. Mol Cell Endocrinol. 2010. PMID: 20638985 Free PMC article.
References
-
- Zhu Y, Xu G, Patel A, McLaughlin MM, Silverman C, Knecht K, Sweitzer S, Li X, McDonnell P, Mirabile R, Zimmerman D, Boyce R, Tierney LA, Hu E, Livi GP, Wolf B, bdel-Meguid SS, Rose GD, Aurora R, Hensley P, Briggs M, Young PR. Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics. 2002;80:144. - PubMed
-
- Cao X, Gao Z, Robert CE, Greene S, Xu G, Xu W, Bell E, Campbell D, Zhu Y, Young R, Trucco M, Markmann JF, Naji A, Wolf BA. Pancreatic-derived factor (FAM3B), a novel islet cytokine, induces apoptosis of insulin-secreting beta-cells. Diabetes. 2003;52:2296. - PubMed
-
- Burkhardt BR, Yang MC, Robert CE, Greene SR, McFadden KK, Yang J, Wu J, Gao Z, Wolf BA. Tissue-specific and glucose-responsive expression of the pancreatic derived factor (PANDER) promoter. Biochim Biophys Acta. 2005;1730:215. - PubMed
-
- Yang J, Robert CE, Burkhardt BR, Young RA, Wu J, Gao Z, Wolf BA. Mechanisms of glucose-induced secretion of pancreatic-derived factor (PANDER or FAM3B) in pancreatic beta-cells. Diabetes. 2005;54:3217. - PubMed
-
- Wang O, Cai K, Pang S, Wang T, Qi D, Zhu Q, Ni Z, Le Y. Mechanisms of glucose-induced expression of pancreatic-derived factor in pancreatic {beta}-cells. Endocrinology. 2007 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

