Discovery of cellular regulation by protein degradation
- PMID: 18708349
- PMCID: PMC3259866
- DOI: 10.1074/jbc.X800009200
Discovery of cellular regulation by protein degradation
Figures
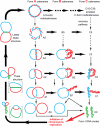
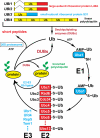

References
-
- Varshavsky, A., Sundin, O., and Bohn, M. (1979) Cell 16 453–466 - PubMed
-
- Sundin, O., and Varshavsky, A. (1980) Cell 21 103–114 - PubMed
-
- Sundin, O., and Varshavsky, A. (1981) Cell 25 659–669 - PubMed
-
- Varshavsky, A., Levinger, L., Sundin, O., Barsoum, J., Özkaynak, E., Swerdlow, P., and Finley, D. (1983) Cold Spring Harb. Symp. Quant. Biol. 47 511–528 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

