Coevolution of function and the folding landscape: correlation with density of native contacts
- PMID: 18708465
- PMCID: PMC2567933
- DOI: 10.1529/biophysj.108.143388
Coevolution of function and the folding landscape: correlation with density of native contacts
Abstract
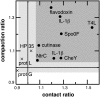
The relationship between the folding landscape and function of evolved proteins is explored by comparison of the folding mechanisms for members of the flavodoxin fold. CheY, Spo0F, and NtrC have unrelated functions and low sequence homology but share an identical topology. Recent coarse-grained simulations show that their folding landscapes are uniquely tuned to properly suit their respective biological functions. Enhanced packing in Spo0F and its limited conformational dynamics compared to CheY or NtrC lead to frustration in its folding landscape. Simulation as well as experimental results correlate with the local density of native contacts for these and a sample of other proteins. In particular, protein regions of low contact density are observed to become structured late in folding; concomitantly, these dynamic regions are often involved in binding or conformational rearrangements of functional importance. These observations help to explain the widespread success of Gō-like coarse-grained models in reproducing protein dynamics.
Figures
Similar articles
-
Insights from coarse-grained Gō models for protein folding and dynamics.Int J Mol Sci. 2009 Mar;10(3):889-905. doi: 10.3390/ijms10030889. Epub 2009 Mar 2. Int J Mol Sci. 2009. PMID: 19399227 Free PMC article. Review.
-
Topological frustration in beta alpha-repeat proteins: sequence diversity modulates the conserved folding mechanisms of alpha/beta/alpha sandwich proteins.J Mol Biol. 2010 Apr 30;398(2):332-50. doi: 10.1016/j.jmb.2010.03.001. Epub 2010 Mar 11. J Mol Biol. 2010. PMID: 20226790 Free PMC article.
-
Folding of proteins with a flavodoxin-like architecture.FEBS J. 2017 Oct;284(19):3145-3167. doi: 10.1111/febs.14077. Epub 2017 Apr 28. FEBS J. 2017. PMID: 28380286 Review.
-
Balancing bond, nonbond, and gō-like terms in coarse grain simulations of conformational dynamics.Methods Mol Biol. 2014;1084:123-40. doi: 10.1007/978-1-62703-658-0_7. Methods Mol Biol. 2014. PMID: 24061919
-
Subdomain competition, cooperativity, and topological frustration in the folding of CheY.J Mol Biol. 2008 Oct 3;382(2):485-95. doi: 10.1016/j.jmb.2008.07.007. Epub 2008 Jul 11. J Mol Biol. 2008. PMID: 18644380 Free PMC article.
Cited by
-
Characterization of the cofactor-induced folding mechanism of a zinc-binding peptide using computationally designed mutants.J Mol Biol. 2009 May 29;389(1):90-102. doi: 10.1016/j.jmb.2009.03.074. Epub 2009 Apr 8. J Mol Biol. 2009. PMID: 19361525 Free PMC article.
-
Energetic frustrations in protein folding at residue resolution: a homologous simulation study of Im9 proteins.PLoS One. 2014 Jan 31;9(1):e87719. doi: 10.1371/journal.pone.0087719. eCollection 2014. PLoS One. 2014. PMID: 24498176 Free PMC article.
-
MD Simulations of tRNA and Aminoacyl-tRNA Synthetases: Dynamics, Folding, Binding, and Allostery.Int J Mol Sci. 2015 Jul 13;16(7):15872-902. doi: 10.3390/ijms160715872. Int J Mol Sci. 2015. PMID: 26184179 Free PMC article. Review.
-
Insights from coarse-grained Gō models for protein folding and dynamics.Int J Mol Sci. 2009 Mar;10(3):889-905. doi: 10.3390/ijms10030889. Epub 2009 Mar 2. Int J Mol Sci. 2009. PMID: 19399227 Free PMC article. Review.
-
Unfolding simulations reveal the mechanism of extreme unfolding cooperativity in the kinetically stable alpha-lytic protease.PLoS Comput Biol. 2010 Feb 26;6(2):e1000689. doi: 10.1371/journal.pcbi.1000689. PLoS Comput Biol. 2010. PMID: 20195497 Free PMC article.
References
-
- Onuchic, J. N., and P. G. Wolynes. 2004. Theory of protein folding. Curr. Opin. Struct. Biol. 14:70–75. - PubMed
-
- Clementi, C., H. Nymeyer, and J. N. Onuchic. 2000. Topological and energetic factors: what determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J. Mol. Biol. 298:937–953. - PubMed
-
- Karanicolas, J., and C. L. Brooks III. 2003. Improved Gō-like models demonstrate the robustness of protein folding mechanisms towards non-native interactions. J. Mol. Biol. 334:309–325. - PubMed
-
- Lee, S. Y., Y. Fujitsuka, D. H. Kim, and S. Takada. 2004. Roles of physical interactions in determining protein folding mechanisms: molecular simulation of protein G and α-spectrin SH3. Proteins. 55:128–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

