Coevolution of function and the folding landscape: correlation with density of native contacts
- PMID: 18708465
- PMCID: PMC2567933
- DOI: 10.1529/biophysj.108.143388
Coevolution of function and the folding landscape: correlation with density of native contacts
Abstract
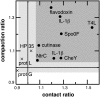
The relationship between the folding landscape and function of evolved proteins is explored by comparison of the folding mechanisms for members of the flavodoxin fold. CheY, Spo0F, and NtrC have unrelated functions and low sequence homology but share an identical topology. Recent coarse-grained simulations show that their folding landscapes are uniquely tuned to properly suit their respective biological functions. Enhanced packing in Spo0F and its limited conformational dynamics compared to CheY or NtrC lead to frustration in its folding landscape. Simulation as well as experimental results correlate with the local density of native contacts for these and a sample of other proteins. In particular, protein regions of low contact density are observed to become structured late in folding; concomitantly, these dynamic regions are often involved in binding or conformational rearrangements of functional importance. These observations help to explain the widespread success of Gō-like coarse-grained models in reproducing protein dynamics.
Figures
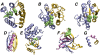
References
-
- Onuchic, J. N., and P. G. Wolynes. 2004. Theory of protein folding. Curr. Opin. Struct. Biol. 14:70–75. - PubMed
-
- Clementi, C., H. Nymeyer, and J. N. Onuchic. 2000. Topological and energetic factors: what determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J. Mol. Biol. 298:937–953. - PubMed
-
- Karanicolas, J., and C. L. Brooks III. 2003. Improved Gō-like models demonstrate the robustness of protein folding mechanisms towards non-native interactions. J. Mol. Biol. 334:309–325. - PubMed
-
- Lee, S. Y., Y. Fujitsuka, D. H. Kim, and S. Takada. 2004. Roles of physical interactions in determining protein folding mechanisms: molecular simulation of protein G and α-spectrin SH3. Proteins. 55:128–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

