O2- and NO-sensing mechanism through the DevSR two-component system in Mycobacterium smegmatis
- PMID: 18708494
- PMCID: PMC2566196
- DOI: 10.1128/JB.00401-08
O2- and NO-sensing mechanism through the DevSR two-component system in Mycobacterium smegmatis
Abstract
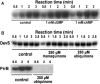
The DevS histidine kinase of Mycobacterium smegmatis contains tandem GAF domains (GAF-A and GAF-B) in its N-terminal sensory domain. The heme iron of DevS is in the ferrous state when purified and is resistant to autooxidation from a ferrous to a ferric state in the presence of O(2). The redox property of the heme and the results of sequence comparison analysis indicate that DevS of M. smegmatis is more closely related to DosT of Mycobacterium tuberculosis than DevS of M. tuberculosis. The binding of O(2) to the deoxyferrous heme led to a decrease in the autokinase activity of DevS, whereas NO binding did not. The regulation of DevS autokinase activity in response to O(2) and NO was not observed in the DevS derivatives lacking its heme, indicating that the ligand-binding state of the heme plays an important role in the regulation of DevS kinase activity. The redox state of the quinone/quinol pool of the respiratory electron transport chain appears not to be implicated in the regulation of DevS activity. Neither cyclic GMP (cGMP) nor cAMP affected DevS autokinase activity, excluding the possibility that the cyclic nucleotides serve as the effector molecules to modulate DevS kinase activity. The three-dimensional structure of the putative GAF-B domain revealed that it has a GAF folding structure without cyclic nucleotide binding capacity.
Figures



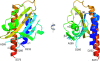
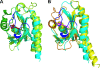


References
-
- Aly, S., K. Wagner, C. Keller, S. Malm, A. Malzan, S. Brandau, F. C. Bange, and S. Ehlers. 2006. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J. Pathol. 210298-305. - PubMed
-
- Aravind, L., and C. P. Ponting. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22458-459. - PubMed
-
- Berman, H., K. Henrick, and H. Nakamura. 2003. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 10980. - PubMed
-
- Charbonneau, H., R. K. Prusti, H. LeTrong, W. K. Sonnenburg, P. J. Mullaney, K. A. Walsh, and J. A. Beavo. 1990. Identification of a noncatalytic cGMP-binding domain conserved in both the cGMP-stimulated and photoreceptor cyclic nucleotide phosphodiesterases. Proc. Natl. Acad. Sci. USA 87288-292. - PMC - PubMed
-
- Dasgupta, N., V. Kapur, K. K. Singh, T. K. Das, S. Sachdeva, K. Jyothisri, and J. S. Tyagi. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuberc. Lung Dis. 80141-159. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

