Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes
- PMID: 18708579
- PMCID: PMC2518813
- DOI: 10.1101/gad.1675908
Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes
Abstract
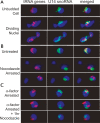
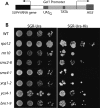
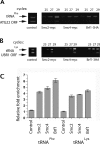
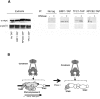
The 274 tRNA genes in Saccharomyces cerevisiae are scattered throughout the linear maps of the 16 chromosomes, but the genes are clustered at the nucleolus when compacted in the nucleus. This clustering is dependent on intact nucleolar organization and contributes to tRNA gene-mediated (tgm) silencing of RNA polymerase II transcription near tRNA genes. After examination of the localization mechanism, we find that the chromosome-condensing complex, condensin, is involved in the clustering of tRNA genes. Conditionally defective mutations in all five subunits of condensin, which we confirm is bound to active tRNA genes in the yeast genome, lead to loss of both pol II transcriptional silencing near tRNA genes and nucleolar clustering of the genes. Furthermore, we show that condensin physically associates with a subcomplex of RNA polymerase III transcription factors on the tRNA genes. Clustering of tRNA genes by condensin appears to be a separate mechanism from their nucleolar localization, as microtubule disruption releases tRNA gene clusters from the nucleolus, but does not disperse the clusters. These observations suggest a widespread role for condensin in gene organization and packaging of the interphase yeast nucleus.
Figures

References
-
- Bartova E., Kozubek S. Nuclear architecture in the light of gene expression and cell differentiation studies. Biol. Cell. 2006;98:323–336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
