Identification of cis-acting sites for condensin loading onto budding yeast chromosomes
- PMID: 18708580
- PMCID: PMC2518811
- DOI: 10.1101/gad.1675708
Identification of cis-acting sites for condensin loading onto budding yeast chromosomes
Abstract
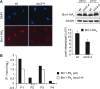
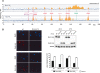
Eukaryotic chromosomes reach their stable rod-shaped appearance in mitosis in a reaction dependent on the evolutionarily conserved condensin complex. Little is known about how and where condensin associates with chromosomes. Here, we analyze condensin binding to budding yeast chromosomes using high-resolution oligonucleotide tiling arrays. Condensin-binding sites coincide with those of the loading factor Scc2/4 of the related cohesin complex. The sites map to tRNA and other genes bound by the RNA polymerase III transcription factor TFIIIC, and ribosomal protein and SNR genes. An ectopic B-box element, recognized by TFIIIC, constitutes a minimal condensin-binding site, and TFIIIC and the Scc2/4 complex promote functional condensin association with chromosomes. A similar pattern of condensin binding is conserved along fission yeast chromosomes. This reveals that TFIIIC-binding sites, including tRNA genes, constitute a hitherto unknown chromosomal feature with important implications for chromosome architecture during both interphase and mitosis.
Figures





References
-
- Aono N., Sutani T., Tomonaga T., Mochida S., Yanagida M. Cnd2 has dual roles in mitotic condensation and interphase. Nature. 2002;417:197–202. - PubMed
-
- Arumugam P., Gruber S., Tanaka K., Haering C.H., Mechtler K., Nasmyth K. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 2003;13:1941–1953. - PubMed
-
- Bazett-Jones D.P., Kimura K., Hirano T. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell. 2002;9:1183–1190. - PubMed
-
- Betts Lindroos H., Ström L., Itoh T., Katou Y., Shirahige K., Sjögren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006;22:755–767. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
