Recognition of misfolded proteins by Lon, a AAA(+) protease
- PMID: 18708584
- PMCID: PMC2518814
- DOI: 10.1101/gad.1670908
Recognition of misfolded proteins by Lon, a AAA(+) protease
Abstract
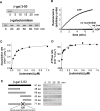
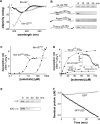
Proteins unfold constantly in cells, especially under stress conditions. Degradation of denatured polypeptides by Lon and related ATP-dependent AAA(+) proteases helps prevent toxic aggregates formation and other deleterious consequences, but how these destructive enzymatic machines distinguish between damaged and properly folded proteins is poorly understood. Here, we show that Escherichia coli Lon recognizes specific sequences -- rich in aromatic residues -- that are accessible in unfolded polypeptides but hidden in most native structures. Denatured polypeptides lacking such sequences are poor substrates. Lon also unfolds and degrades stably folded proteins with accessible recognition tags. Thus, protein architecture and the positioning of appropriate targeting sequences allow Lon degradation to be dependent or independent of the folding status of a protein. Our results suggest that Lon can recognize multiple signals in unfolded polypeptides synergistically, resulting in nanomolar binding and a mechanism for discriminating irreversibly damaged proteins from transiently unfolded elements of structure.
Figures
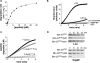


References
-
- Baldwin E.P., Matthews B.W. Core-packing constraints, hydrophobicity and protein design. Curr. Opin. Biotechnol. 1994;5:396–402. - PubMed
-
- Bukau B., Weissman J., Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. - PubMed
-
- Burton R.E., Baker T.A., Sauer R.T. Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat. Struct. Mol. Biol. 2005;12:245–251. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
