Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development
- PMID: 18710561
- PMCID: PMC2575520
- DOI: 10.1186/gb-2008-9-8-r130
Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development
Abstract
Background: As nonmotile organisms, plants must rapidly adapt to ever-changing environmental conditions, including those caused by daily light/dark cycles. One important mechanism for anticipating and preparing for such predictable changes is the circadian clock. Nearly all organisms have circadian oscillators that, when they are in phase with the Earth's rotation, provide a competitive advantage. In order to understand how circadian clocks benefit plants, it is necessary to identify the pathways and processes that are clock controlled.
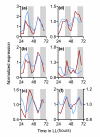
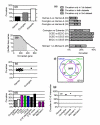
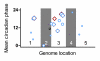
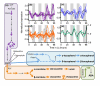
Results: We have integrated information from multiple circadian microarray experiments performed on Arabidopsis thaliana in order to better estimate the fraction of the plant transcriptome that is circadian regulated. Analyzing the promoters of clock-controlled genes, we identified circadian clock regulatory elements correlated with phase-specific transcript accumulation. We have also identified several physiological pathways enriched for clock-regulated changes in transcript abundance, suggesting they may be modulated by the circadian clock.
Conclusion: Our analysis suggests that transcript abundance of roughly one-third of expressed A. thaliana genes is circadian regulated. We found four promoter elements, enriched in the promoters of genes with four discrete phases, which may contribute to the time-of-day specific changes in the transcript abundance of these genes. Clock-regulated genes are over-represented among all of the classical plant hormone and multiple stress response pathways, suggesting that all of these pathways are influenced by the circadian clock. Further exploration of the links between the clock and these pathways will lead to a better understanding of how the circadian clock affects plant growth and leads to improved fitness.
Figures

References
-
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. - PubMed
-
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–1486. - PubMed
-
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

