Genome sequence of the Bacteroides fragilis phage ATCC 51477-B1
- PMID: 18710568
- PMCID: PMC2535602
- DOI: 10.1186/1743-422X-5-97
Genome sequence of the Bacteroides fragilis phage ATCC 51477-B1
Abstract
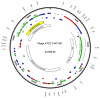
The genome of a fecal pollution indicator phage, Bacteroides fragilis ATCC 51477-B1, was sequenced and consisted of 44,929 bases with a G+C content of 38.7%. Forty-six putative open reading frames were identified and genes were organized into functional clusters for host specificity, lysis, replication and regulation, and packaging and structural proteins.
Figures

References
-
- Dick LK, Bernhard AE, Brodeur TJ, Domingo JWS, Simpson JM, Walters SP, Field KG. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Applied and Environmental Microbiology. 2005;71:3184–3191. doi: 10.1128/AEM.71.6.3184-3191.2005. - DOI - PMC - PubMed
-
- Puig A, Jofre J, Araujo R. Bacteriophages infecting various Bacteroides fragilis strains differ in their capacity to distinguish human from animal faecal pollution. Water Science and Technology. 1997;35:359–362. doi: 10.1016/S0273-1223(97)00285-0. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

