Factors in tissue handling and processing that impact RNA obtained from formalin-fixed, paraffin-embedded tissue
- PMID: 18711211
- PMCID: PMC2569903
- DOI: 10.1369/jhc.2008.951863
Factors in tissue handling and processing that impact RNA obtained from formalin-fixed, paraffin-embedded tissue
Abstract
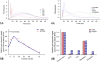
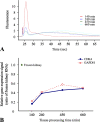
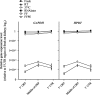
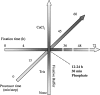
Formalin-fixed, paraffin-embedded (FFPE) tissue is the most common specimen available for molecular assays on tissue after diagnostic histopathological examination. RNA from FFPE tissue suffers from strand breakage and cross-linking. Despite excellent extraction methods, RNA quality from FFPE material remains variable. To address the RNA quality factors within FFPE tissues, we studied RNA quality, isolating individual elements of the tissue fixation and processing including length of fixation in formalin and the type of buffer incorporated in the fixative. We examined the impact of the length of the tissue processing cycle as well. The optimal fixation period of 12-24 hr in phosphate-buffered formalin resulted in better-quality RNA. Longer tissue processing times were associated with higher quality RNA. We determined that the middle region of gene suffers less damage by these processes as shown by real-time quantitative RT-PCR. These data provide key information for the development of methods of analysis of gene expression in archival FFPE tissues and contribute to the establishment of objective standards for the processing and handling of tissue in surgical pathology. This manuscript contains online supplemental material at http://www.jhc.org. Please visit this article online to view these materials.
Figures



References
-
- Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Komacker K (2003) Chipping away at the chip bias: RNA degradation in microarray analysis. Nat Genet 35:292–293 - PubMed
-
- Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, et al. (2006) Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol 24:1115–1122 - PubMed
-
- Chung JY, Braunschweig T, Hewitt SM (2006) Optimization of recovery of RNA from formalin-fixed paraffin-embedded tissue. Diagn Mol Pathol 15:229–236 - PubMed
-
- Coolidge BJ, Howard RM (1979) Animal Histology Procedures (NIH Publication 80–275). Bethesda, National Institutes of Health
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

