Using expression profiles of Caenorhabditis elegans neurons to identify genes that mediate synaptic connectivity
- PMID: 18711638
- PMCID: PMC2517614
- DOI: 10.1371/journal.pcbi.1000120
Using expression profiles of Caenorhabditis elegans neurons to identify genes that mediate synaptic connectivity
Abstract
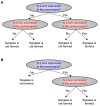
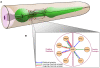
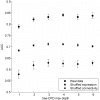
Synaptic wiring of neurons in Caenorhabditis elegans is largely invariable between animals. It has been suggested that this feature stems from genetically encoded molecular markers that guide the neurons in the final stage of synaptic formation. Identifying these markers and unraveling the logic by which they direct synapse formation is a key challenge. Here, we address this task by constructing a probabilistic model that attempts to explain the neuronal connectivity diagram of C. elegans as a function of the expression patterns of its neurons. By only considering neuron pairs that are known to be connected by chemical or electrical synapses, we focus on the final stage of synapse formation, in which neurons identify their designated partners. Our results show that for many neurons the neuronal expression map of C. elegans can be used to accurately predict the subset of adjacent neurons that will be chosen as its postsynaptic partners. Notably, these predictions can be achieved using the expression patterns of only a small number of specific genes that interact in a combinatorial fashion.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Durbin RM. Cambridge, UK: Cambridge University; 1987. Studies on the Development and Organisation of the Nervous System of Caenorhabditis elegans [PhD dissertation].
-
- White JG. Neuronal connectivity in Caenorhabditis elegans. Trends Neurosci. 1985;8:277–283.
-
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. - PubMed
-
- Sperry RW. Effect of 180 degree rotation of the retinal field on visuomotor coordination. J Exp Zool. 1943;92:263–279.
MeSH terms
LinkOut - more resources
Full Text Sources

