Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis
- PMID: 18713001
- PMCID: PMC2650809
- DOI: 10.1515/BC.2008.130
Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis
Abstract
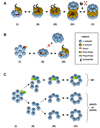
The 26S proteasome is the key eukaryotic protease responsible for the degradation of intracellular proteins. Protein degradation by the 26S proteasome plays important roles in numerous cellular processes, including the cell cycle, differentiation, apoptosis, and the removal of damaged or misfolded proteins. How this 2.5-MDa complex, composed of at least 32 different polypeptides, is assembled in the first place is not well understood. However, it has become evident that this complicated task is facilitated by a framework of protein factors that chaperone the nascent proteasome through its various stages of assembly. We review here the known proteasome-specific assembly factors, most only recently discovered, and describe their potential roles in proteasome assembly, with an emphasis on the many remaining unanswered questions about this intricate process of assisted self-assembly.
Figures
References
-
- Bahar R, J OW, Kawamura K, Seimiya M, Wang Y, Hatano M, Okada S, Tokuhisa T, Watanabe T, Tagawa M. Growth retardation, polyploidy, and multinucleation induced by Clast3, a novel cell cycle-regulated protein. J. Biol. Chem. 2002;277:40012–40019. - PubMed
-
- Brennan RJ, Schiestl RH. Cadmium is an inducer of oxidative stress in yeast. Mutat. Res. 1996;356:171–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
