The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes
- PMID: 18713319
- PMCID: PMC4623567
- DOI: 10.1111/j.1365-2958.2008.06408.x
The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes
Abstract
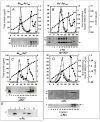
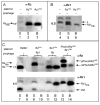
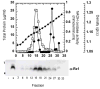
Bacteriophage lambda has four adjacent genes -S, R, Rz and Rz1- dedicated to host cell lysis. While S, encoding the holin and antiholin, and R, encoding the endolysin, have been intensively studied, the products of Rz and Rz1 have not been characterized at either the structural or functional levels. Rz1 is an outer membrane lipoprotein and our results indicate that Rz is a type II signal anchor protein. Here we present evidence that an Rz-Rz1 complex that spans the periplasm carries out the final step in the process of host lysis. These results are discussed in terms of a model where endolysin-mediated degradation of the cell wall is a prerequisite for conformational changes in the Rz-Rz1 complex leading to the juxtaposition and fusion of the IM and OM. Fusion of the two membranes removes the last physical barrier to efficient release of progeny virions.
Figures


References
-
- Bartel PL, Roecklein JA, SenGupta D, Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nat Genet. 1996;12:72–77. - PubMed
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. - PubMed
-
- Bernhardt TG, Roof WD, Young R. The Escherichia coli FKBP-type PPIase SlyD is required for the stabilization of the E lysis protein of bacteriophage ϕX174. Mol Microbiol. 2002;45:99–108. - PubMed
-
- Bienkowska-Szewczyk K, Taylor A. Murein transglycosylase from phage λ lysate: purification and properties. Biochim Biophys Acta. 1980;615:489–496. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

