Enhanced protein expression in the baculovirus/insect cell system using engineered SUMO fusions
- PMID: 18713650
- PMCID: PMC2585507
- DOI: 10.1016/j.pep.2008.07.010
Enhanced protein expression in the baculovirus/insect cell system using engineered SUMO fusions
Abstract
Recombinant protein expression in insect cells varies greatly from protein to protein. A fusion tag that is not only a tool for detection and purification, but also enhances expression and/or solubility would greatly facilitate both structure/function studies and therapeutic protein production. We have shown that fusion of SUMO (small ubiquitin-related modifier) to several test proteins leads to enhanced expression levels in Escherichia coli. In eukaryotic expression systems, however, the SUMO tag could be cleaved by endogenous desumoylase. In order to adapt SUMO-fusion technology to these systems, we have developed an alternative SUMO-derived tag, designated SUMOstar, which is not processed by native SUMO proteases. In the present study, we tested the SUMOstar tag in a baculovirus/insect cell system with several proteins, i.e. mouse UBP43, human tryptase beta II, USP4, USP15, and GFP. Our results demonstrate that fusion to SUMOstar enhanced protein expression levels at least 4-fold compared to either the native or His(6)-tagged proteins. We isolated active SUMOstar tagged UBP43, USP4, USP15, and GFP. Tryptase was active following cleavage with a SUMOstar specific protease. The SUMOstar system will make significant impact in difficult-to-express proteins and especially to those proteins that require the native N-terminal residue for function.
Figures



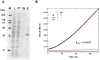
 ) Reaction mixture with heparin and SUMOstar-tryptase but without SUMOstar protease. (
) Reaction mixture with heparin and SUMOstar-tryptase but without SUMOstar protease. (
 ) Reaction mixture minus SUMOstar-tryptase and SUMOstar protease. In a separate experiment SUMOstar protease was found to have no activity against the tryptase substrate (data not shown.)
) Reaction mixture minus SUMOstar-tryptase and SUMOstar protease. In a separate experiment SUMOstar protease was found to have no activity against the tryptase substrate (data not shown.)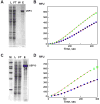
 ) or 10 (
) or 10 (
 ) μL of the concentrated pool of USP4. (D) Ub-PLA2 processing activity of either 5 (
) μL of the concentrated pool of USP4. (D) Ub-PLA2 processing activity of either 5 (
 ) or 10 (
) or 10 (
 ) μL of the concentrated pool of USP15. For both, the line represents the fit of the data to equation 1. Boxed points were excluded from the calculation due to depletion of the PLA2 substrate.
) μL of the concentrated pool of USP15. For both, the line represents the fit of the data to equation 1. Boxed points were excluded from the calculation due to depletion of the PLA2 substrate.Similar articles
-
Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO.Protein Sci. 2006 Jan;15(1):182-9. doi: 10.1110/ps.051812706. Epub 2005 Dec 1. Protein Sci. 2006. PMID: 16322573 Free PMC article.
-
SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems.Methods Mol Biol. 2009;497:303-17. doi: 10.1007/978-1-59745-566-4_20. Methods Mol Biol. 2009. PMID: 19107426 Review.
-
Human SUMO fusion systems enhance protein expression and solubility.Protein Expr Purif. 2010 Oct;73(2):203-8. doi: 10.1016/j.pep.2010.05.001. Epub 2010 May 10. Protein Expr Purif. 2010. PMID: 20457256
-
Expression and immunoaffinity purification of recombinant dengue virus 2 NS1 protein as a cleavable SUMOstar fusion.Protein Expr Purif. 2012 Mar;82(1):20-5. doi: 10.1016/j.pep.2011.11.003. Epub 2011 Nov 11. Protein Expr Purif. 2012. PMID: 22100526
-
SUMO fusion technology for enhanced protein expression and purification in prokaryotes and eukaryotes.Methods Mol Biol. 2011;705:15-30. doi: 10.1007/978-1-61737-967-3_2. Methods Mol Biol. 2011. PMID: 21125378 Review.
Cited by
-
The Xenopus laevis Atg4B Protease: Insights into Substrate Recognition and Application for Tag Removal from Proteins Expressed in Pro- and Eukaryotic Hosts.PLoS One. 2015 Apr 29;10(4):e0125099. doi: 10.1371/journal.pone.0125099. eCollection 2015. PLoS One. 2015. PMID: 25923686 Free PMC article.
-
Biosynthesis of medicinal tropane alkaloids in yeast.Nature. 2020 Sep;585(7826):614-619. doi: 10.1038/s41586-020-2650-9. Epub 2020 Sep 2. Nature. 2020. PMID: 32879484 Free PMC article.
-
Peptide Tags and Domains for Expression and Detection of Mammalian Membrane Proteins at the Cell Surface.Methods Mol Biol. 2022;2507:337-358. doi: 10.1007/978-1-0716-2368-8_18. Methods Mol Biol. 2022. PMID: 35773591 Free PMC article.
-
Xpo7 is a broad-spectrum exportin and a nuclear import receptor.J Cell Biol. 2018 Jul 2;217(7):2329-2340. doi: 10.1083/jcb.201712013. Epub 2018 May 10. J Cell Biol. 2018. PMID: 29748336 Free PMC article.
-
Doa10 is a membrane protein retrotranslocase in ER-associated protein degradation.Elife. 2020 Jun 26;9:e56945. doi: 10.7554/eLife.56945. Elife. 2020. PMID: 32588820 Free PMC article.
References
-
- Hochstrasser M. SP-RING for SUMO: New Functions Bloom for a Ubiquitin-like Protein. Cell. 2001;107:5–8. - PubMed
-
- Johnson ES. Protein modification by SUMO. Ann Rev Biochem. 2004;73:355–82. - PubMed
-
- Kim KI, Baek SH, Chung CH. Versatile protein tag, SUMO: its enzymology and biological function. J Cell Physiol. 2002;191:257–68. - PubMed
-
- Lalioti VS, Vergarajauregui S, Pulido D, Sandoval IV. The Insulin-sensitive Glucose Transporter, GLUT4, Interacts Physically with Daxx. TWO PROTEINS WITH CAPACITY TO BIND Ubc9 AND CONJUGATED TO SUMO1. J Biol Chem. 2002;277:19783–19791. - PubMed
-
- Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

