Ecto-nucleoside triphosphate diphosphohydrolase 1 (E-NTPDase1/CD39) regulates neutrophil chemotaxis by hydrolyzing released ATP to adenosine
- PMID: 18713747
- PMCID: PMC2568933
- DOI: 10.1074/jbc.M800039200
Ecto-nucleoside triphosphate diphosphohydrolase 1 (E-NTPDase1/CD39) regulates neutrophil chemotaxis by hydrolyzing released ATP to adenosine
Abstract
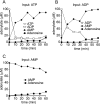
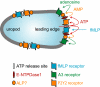
Polymorphonuclear neutrophils release ATP in response to stimulation by chemoattractants, such as the peptide N-formyl-methionyl-leucyl-phenylalanine. Released ATP and the hydrolytic product adenosine regulate chemotaxis of neutrophils by sequentially activating purinergic nucleotide and adenosine receptors, respectively. Here we show that that ecto-nucleoside triphosphate diphosphohydrolase 1 (E-NTPDase1, CD39) is a critical enzyme for hydrolysis of released ATP by neutrophils and for cell migration in response to multiple agonists (N-formyl-methionyl-leucyl-phenylalanine, interleukin-8, and C5a). Upon stimulation of human neutrophils or differentiated HL-60 cells in a chemotactic gradient, E-NTPDase1 tightly associates with the leading edge of polarized cells during chemotaxis. Inhibition of E-NTPDase1 reduces the migration speed of neutrophils but not their ability to detect the orientation of the gradient field. Studies of neutrophils from E-NTPDase1 knock-out mice reveal similar impairments of chemotaxis in vitro and in vivo. Thus, E-NTPDase1 plays an important role in regulating neutrophil chemotaxis by facilitating the hydrolysis of extracellular ATP.
Figures


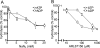



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

