Lifelong reduction of LDL-cholesterol related to a common variant in the LDL-receptor gene decreases the risk of coronary artery disease--a Mendelian Randomisation study
- PMID: 18714375
- PMCID: PMC2500189
- DOI: 10.1371/journal.pone.0002986
Lifelong reduction of LDL-cholesterol related to a common variant in the LDL-receptor gene decreases the risk of coronary artery disease--a Mendelian Randomisation study
Erratum in
- PLoS ONE. 2008;3(9). doi: 10.1371/annotation/9f64c41a-8cf6-40f2-8988-0d48b04dd8cb. Schrezenmeier, Jürgen [corrected to Schrezenmeir, Jürgen]
Abstract
Background: Rare mutations of the low-density lipoprotein receptor gene (LDLR) cause familial hypercholesterolemia, which increases the risk for coronary artery disease (CAD). Less is known about the implications of common genetic variation in the LDLR gene regarding the variability of cholesterol levels and risk of CAD.
Methods: Imputed genotype data at the LDLR locus on 1 644 individuals of a population-based sample were explored for association with LDL-C level. Replication of association with LDL-C level was sought for the most significant single nucleotide polymorphism (SNP) within the LDLR gene in three European samples comprising 6 642 adults and 533 children. Association of this SNP with CAD was examined in six case-control studies involving more than 15 000 individuals.
Findings: Each copy of the minor T allele of SNP rs2228671 within LDLR (frequency 11%) was related to a decrease of LDL-C levels by 0.19 mmol/L (95% confidence interval (CI) [0.13-0.24] mmol/L, p = 1.5x10(-10)). This association with LDL-C was uniformly found in children, men, and women of all samples studied. In parallel, the T allele of rs2228671 was associated with a significantly lower risk of CAD (Odds Ratio per copy of the T allele: 0.82, 95% CI [0.76-0.89], p = 2.1x10(-7)). Adjustment for LDL-C levels by logistic regression or Mendelian Randomisation models abolished the significant association between rs2228671 with CAD completely, indicating a functional link between the genetic variant at the LDLR gene locus, change in LDL-C and risk of CAD.
Conclusion: A common variant at the LDLR gene locus affects LDL-C levels and, thereby, the risk for CAD.
Conflict of interest statement
Figures




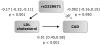
References
-
- Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr, Clark LT, et al. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Circulation. 2004;110:227–239. - PubMed
-
- Brown MS, Goldstein JL. Biomedicine. Lowering LDL–not only how low, but how long? Science. 2006;311:1721–1723. - PubMed
-
- Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. - PubMed
-
- Brown MS, Goldstein JL. Expression of the familial hypercholesterolemia gene in heterozygotes: mechanism for a dominant disorder in man. Science. 1974;185:61–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

