The cellular prion protein PrP(c) is involved in the proliferation of epithelial cells and in the distribution of junction-associated proteins
- PMID: 18714380
- PMCID: PMC2500194
- DOI: 10.1371/journal.pone.0003000
The cellular prion protein PrP(c) is involved in the proliferation of epithelial cells and in the distribution of junction-associated proteins
Erratum in
- PLoS ONE. 2008;3(9). doi: 10.1371/annotation/0b364095-9f93-4cb9-9a2e-aae5ed1bf362
- PLoS ONE. 2008;3(9). doi: 10.1371/annotation/e5a42567-afa3-422a-886e-44ca642c6fe2. Thievend, Cathy Pichol [corrected to Pichol Thievend, Cathy]
Abstract
Background: The physiological function of the ubiquitous cellular prion protein, PrP(c), is still under debate. It was essentially studied in nervous system, but poorly investigated in epithelial cells. We previously reported that PrP(c) is targeted to cell-cell junctions of polarized epithelial cells, where it interacts with c-Src.
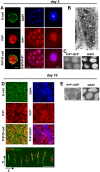
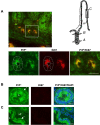
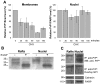
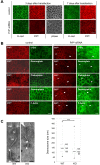
Methodology/findings: We show here that, in cultured human enterocytes and in intestine in vivo, the mature PrP(c) is differentially targeted either to the nucleus in dividing cells or to cell-cell contacts in polarized/differentiated cells. By proteomic analysis, we demonstrate that the junctional PrP(c) interacts with cytoskeleton-associated proteins, such as gamma- and beta-actin, alpha-spectrin, annexin A2, and with the desmosome-associated proteins desmoglein, plakoglobin and desmoplakin. In addition, co-immunoprecipitation experiments revealed complexes associating PrP(c), desmoglein and c-Src in raft domains. Through siRNA strategy, we show that PrP(c) is necessary to complete the process of epithelial cell proliferation and for the sub-cellular distribution of proteins involved in cell architecture and junctions. Moreover, analysis of the architecture of the intestinal epithelium of PrP(c) knock-out mice revealed a net decrease in the size of desmosomal junctions and, without change in the amount of BrdU incorporation, a shortening of the length of intestinal villi.
Conclusions/significance: From these results, PrP(c) could be considered as a new partner involved in the balance between proliferation and polarization/differentiation in epithelial cells.
Conflict of interest statement
Figures

Similar articles
-
Requirement of cellular prion protein for intestinal barrier function and mislocalization in patients with inflammatory bowel disease.Gastroenterology. 2012 Jul;143(1):122-32.e15. doi: 10.1053/j.gastro.2012.03.029. Epub 2012 Mar 22. Gastroenterology. 2012. PMID: 22446194
-
The nucleo-junctional interplay of the cellular prion protein: A new partner in cancer-related signaling pathways?Prion. 2016 Mar 3;10(2):143-52. doi: 10.1080/19336896.2016.1163457. Prion. 2016. PMID: 27216988 Free PMC article.
-
Involvement of the Prion Protein in the Protection of the Human Bronchial Epithelial Barrier Against Oxidative Stress.Antioxid Redox Signal. 2019 Jul 1;31(1):59-74. doi: 10.1089/ars.2018.7500. Epub 2019 Feb 4. Antioxid Redox Signal. 2019. PMID: 30569742
-
New insights into cellular prion protein (PrPc) functions: the "ying and yang" of a relevant protein.Brain Res Rev. 2009 Oct;61(2):170-84. doi: 10.1016/j.brainresrev.2009.06.002. Epub 2009 Jun 10. Brain Res Rev. 2009. PMID: 19523487 Review.
-
Epithelial junctions and polarity: complexes and kinases.Curr Opin Nephrol Hypertens. 2008 Sep;17(5):506-12. doi: 10.1097/MNH.0b013e32830baaae. Curr Opin Nephrol Hypertens. 2008. PMID: 18695392 Free PMC article. Review.
Cited by
-
Common themes in PrP signaling: the Src remains the same.Front Cell Dev Biol. 2014 Oct 28;2:63. doi: 10.3389/fcell.2014.00063. eCollection 2014. Front Cell Dev Biol. 2014. PMID: 25364767 Free PMC article.
-
The Role of Cellular Prion Protein in Cancer Biology: A Potential Therapeutic Target.Front Oncol. 2021 Sep 14;11:742949. doi: 10.3389/fonc.2021.742949. eCollection 2021. Front Oncol. 2021. PMID: 34595121 Free PMC article. Review.
-
PrPC expression and prion seeding activity in the alimentary tract and lymphoid tissue of deer.PLoS One. 2017 Sep 7;12(9):e0183927. doi: 10.1371/journal.pone.0183927. eCollection 2017. PLoS One. 2017. PMID: 28880938 Free PMC article.
-
The prion protein is critical for DNA repair and cell survival after genotoxic stress.Nucleic Acids Res. 2015 Jan;43(2):904-16. doi: 10.1093/nar/gku1342. Epub 2014 Dec 24. Nucleic Acids Res. 2015. PMID: 25539913 Free PMC article.
-
Physiological Functions of the Cellular Prion Protein.Front Mol Biosci. 2017 Apr 6;4:19. doi: 10.3389/fmolb.2017.00019. eCollection 2017. Front Mol Biosci. 2017. PMID: 28428956 Free PMC article. Review.
References
-
- Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell. 1998;93:337–348. - PubMed
-
- Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. - PubMed
-
- Weissmann C, Bueler H, Fischer M, Sailer A, Aguzzi A, et al. PrP-deficient mice are resistant to scrapie. Ann N Y Acad Sci. 1994;724:235–240. - PubMed
-
- Bendheim PE, Brown HR, Rudelli RD, Scala LJ, Goller NL, et al. Nearly ubiquitous tissue distribution of the scrapie agent precursor protein. Neurology. 1992;42:149–156. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

