Insights into the catalytic mechanism of tyrosine phenol-lyase from X-ray structures of quinonoid intermediates
- PMID: 18715865
- PMCID: PMC2662015
- DOI: 10.1074/jbc.M802061200
Insights into the catalytic mechanism of tyrosine phenol-lyase from X-ray structures of quinonoid intermediates
Abstract
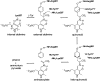
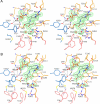
Amino acid transformations catalyzed by a number of pyridoxal 5'-phosphate (PLP)-dependent enzymes involve abstraction of the Calpha proton from an external aldimine formed between a substrate and the cofactor leading to the formation of a quinonoid intermediate. Despite the key role played by the quinonoid intermediates in the catalysis by PLP-dependent enzymes, limited accurate information is available about their structures. We trapped the quinonoid intermediates of Citrobacter freundii tyrosine phenol-lyase with L-alanine and L-methionine in the crystalline state and determined their structures at 1.9- and 1.95-A resolution, respectively, by cryo-crystallography. The data reveal a network of protein-PLP-substrate interactions that stabilize the planar geometry of the quinonoid intermediate. In both structures the protein subunits are found in two conformations, open and closed, uncovering the mechanism by which binding of the substrate and restructuring of the active site during its closure protect the quinonoid intermediate from the solvent and bring catalytically important residues into positions suitable for the abstraction of phenol during the beta-elimination of L-tyrosine. In addition, the structural data indicate a mechanism for alanine racemization involving two bases, Lys-257 and a water molecule. These two bases are connected by a hydrogen bonding system allowing internal transfer of the Calpha proton.
Figures


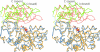



References
-
- Kumagai, H., Yamada, H., Matsui, H., Ohkishi, H., and Ogata, K. (1970) J. Biol. Chem. 245 1767-1772 - PubMed
-
- Phillips, R. S. (1987) Arch. Biochem. Biophys. 256 302-310 - PubMed
-
- Faleev, N. G., Zhukov, Y. N., Khurs, E. N., Gogoleva, O. I., Barbolina, M. V., Bazhulina, N. P., Belikov, V. M., Demidkina, T. V., and Khomutov, R. M. (2000) Eur. J. Biochem. 267 6897-6902 - PubMed
-
- Enei, H., Nakazawa, H., Matsui, H., Okumura, S., and Yamada, H. (1972) FEBS Lett. 21 39-41 - PubMed
-
- Yamada, H., and Kumagai, H. (1975) Adv. Appl. Microbiol. 19 249-288 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

