An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia
- PMID: 18716133
- PMCID: PMC2954679
- DOI: 10.1182/blood-2008-02-134411
An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia
Abstract
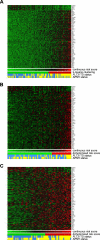
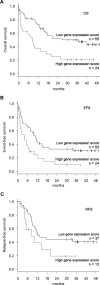
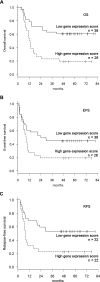
Patients with cytogenetically normal acute myeloid leukemia (CN-AML) show heterogeneous treatment outcomes. We used gene-expression profiling to develop a gene signature that predicts overall survival (OS) in CN-AML. Based on data from 163 patients treated in the German AMLCG 1999 trial and analyzed on oligonucleotide microarrays, we used supervised principal component analysis to identify 86 probe sets (representing 66 different genes), which correlated with OS, and defined a prognostic score based on this signature. When applied to an independent cohort of 79 CN-AML patients, this continuous score remained a significant predictor for OS (hazard ratio [HR], 1.85; P = .002), event-free survival (HR = 1.73; P = .001), and relapse-free survival (HR = 1.76; P = .025). It kept its prognostic value in multivariate analyses adjusting for age, FLT3 ITD, and NPM1 status. In a validation cohort of 64 CN-AML patients treated on CALGB study 9621, the score also predicted OS (HR = 4.11; P < .001), event-free survival (HR = 2.90; P < .001), and relapse-free survival (HR = 3.14, P < .001) and retained its significance in a multivariate model for OS. In summary, we present a novel gene-expression signature that offers additional prognostic information for patients with CN-AML.
Figures

References
-
- Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92:2322–2333. - PubMed
-
- Mrozek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. - PubMed
-
- Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. - PubMed
-
- Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

