Novel SLC7A7 large rearrangements in lysinuric protein intolerance patients involving the same AluY repeat
- PMID: 18716612
- PMCID: PMC2985956
- DOI: 10.1038/ejhg.2008.145
Novel SLC7A7 large rearrangements in lysinuric protein intolerance patients involving the same AluY repeat
Abstract
Lysinuric protein intolerance (LPI) is a rare autosomal inherited disease caused by defective cationic aminoacid transport 4F2hc/y(+)LAT-1 at the basolateral membrane of epithelial cells in the intestine and kidney. LPI is a multisystemic disease with a variety of clinical symptoms such as hepatosplenomegaly, osteoporosis, hypotonia, developmental delay, pulmonary insufficiency or end-stage renal disease. The SLC7A7 gene, which encodes the y(+)LAT-1 protein, is mutated in LPI patients. Mutation analysis of the promoter localized in intron 1 and all exons of the SLC7A7 gene was performed in 11 patients from 9 unrelated LPI families. Point mutation screening was performed by exon direct sequencing and a new multiplex ligation probe amplification (MLPA) assay was set up for large rearrangement analysis. Eleven SLC7A7-specific mutations were identified, seven of them were novel: p.L124P, p.C425R, p.R468X, p.Y274fsX21, c.625+1G>C, DelE4-E11 and DelE6-E11. The novel large deletions originated by the recombination of Alu repeats at introns 3 and 5, respectively, with the same AluY sequence localized at the SLC7A7 3' region. The novel MLPA assay is robust and valuable for LPI molecular diagnosis. Our results suggest that genomic rearrangements of SLC7A7 play a more important role in LPI than has been reported, increasing the detection rate from 5.1 to 21.4%. Moreover, the 3' region AluY repeat could be a recombination hot spot as it is involved in 38% of all SLC7A7 rearranged chromosomes described so far.
Figures

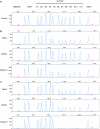

References
-
- Simell O.Lysinuric protein intolerance and other cationic aminoaciduriasin Scriver CR, Beaudet AL, Sly WS, Valle D (eds):The Metabolic and Molecular Bases of Inherited Disease New York, McGraw-Hill; 2001. Vol III, pp4933–4956.
-
- Mykkanen J, Torrents D, Pineda M, et al. Functional analysis of novel mutations in y(+)LAT-1 amino acid transporter gene causing lysinuric protein intolerance (LPI) Hum Mol Genet. 2000;9:431–438. - PubMed
-
- Noguchi A, Shoji Y, Koizumi A, et al. SLC7A7 genomic structure and novel variants in three Japanese lysinuric protein intolerance families. Hum Mutat. 2000;15:367–372. - PubMed
-
- Cimbalistiene L, Lehnert W, Huoponen K, Kucinskas V. First reported case of lysinuric protein intolerance (LPI) in Lithuania, confirmed biochemically and by DNA analysis. J Appl Genet. 2007;48:277–280. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

