Chain dynamics of nascent polypeptides emerging from the ribosome
- PMID: 18717565
- PMCID: PMC2572860
- DOI: 10.1021/cb800059u
Chain dynamics of nascent polypeptides emerging from the ribosome
Abstract
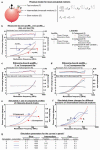
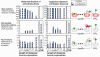
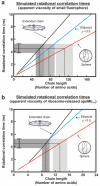
Very little is known about the conformation of polypeptides emerging from the ribosome during protein biosynthesis. Here, we explore the dynamics of ribosome-bound nascent polypeptides and proteins in Escherichia coli by dynamic fluorescence depolarization and assess the population of cotranslationally active chaperones trigger factor (TF) and DnaK. E. coli cell-free technology and fluorophore-linked E. coli Met-tRNA f Met enable selective site-specific labeling of nascent proteins at the N-terminal methionine. For the first time, direct spectroscopic evidence captures the generation of independent nascent chain motions for a single-domain protein emerging from the ribosome (apparent rotational correlation time approximately 5 ns), during the intermediate and late stages of polypeptide elongation. Such motions are detected only for a sequence encoding a globular protein and not for a natively unfolded control, suggesting that the independent nascent chain dynamics may be a signature of folding-competent sequences. In summary, we observe multicomponent, severely rotationally restricted, and strongly chain length/sequence-dependent nascent chain dynamics.
Figures




Comment in
-
Folding on the assembly line.ACS Chem Biol. 2008 Sep 19;3(9):527-9. doi: 10.1021/cb800216n. ACS Chem Biol. 2008. PMID: 18803369
References
-
- Schuwirth BS, Borovinskaya MA, et al. Structures of the bacterial ribosome at 3.5 .ANG. resolution. Science. 2005;310:827–834. - PubMed
-
- Nissen P, Hansen J, et al. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. - PubMed
-
- Harms J, Schluenzen F, et al. High Resolution Structure of the Large Ribosomal Subunit from a Mesophilic Eubacterium. Cell. 2001;107:679–688. - PubMed
-
- Ferbitz L, Maier T, et al. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 2004;431:590–596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

