In vivo and in vitro studies of a functional peroxisome proliferator-activated receptor gamma response element in the mouse pdx-1 promoter
- PMID: 18718916
- PMCID: PMC2583321
- DOI: 10.1074/jbc.M801813200
In vivo and in vitro studies of a functional peroxisome proliferator-activated receptor gamma response element in the mouse pdx-1 promoter
Abstract
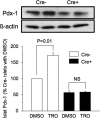
We reported that peroxisome proliferator-activated receptor gamma (PPARgamma) transcriptionally regulates the beta-cell differentiation factor pancreatic duodenal homeobox (PDX)-1 based on in vitro RNA interference studies. We have now studied mice depleted of PPARgamma within the pancreas (PANC PPARgamma(-/-)) created by a Cre/loxP recombinase system, with Cre driven by the pdx-1 promoter. Male PANC PPARgamma(-/-) mice were hyperglycemic at 8 weeks of age (8.1+/-0.2 mM versus 6.4+/-0.3 mM, p=0.009) with islet cytoarchitecture and pancreatic mass of islet beta-cells that were indistinguishable from the controls. Islet PDX-1 mRNA (p=0.001) and protein levels (p=0.003) were lowered 60 and 40%, respectively, in tandem with impaired glucose-induced insulin secretion and loss of thiazolidinedione-induced increase in PDX-1 expression. We next identified a putative PPAR-response element (PPRE) in the mouse pdx-1 promoter with substantial homology to the corresponding region of the human PDX-1 promoter. Electrophoretic mobility supershift assays with nuclear extracts from beta-cell lines and mouse islets, also in vitro translated PPARgamma and retinoid X receptor, and chromatin immunoprecipitation analysis demonstrated specific binding of PPARgamma and retinoid X receptor to the human and mouse pdx-1 x PPREs. Transient transfection assays of beta-cells with reporter constructs of mutated PPREs showed dramatically reduced pdx-1 promoter activity. In summary, we have presented in vivo and in vitro evidence showing PPARgamma regulation of pdx-1 transcription in beta-cells, plus our results support an important regulatory role for PPARgamma in beta-cell physiology and thiazolidinedione pharmacology of type 2 diabetes.
Figures


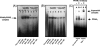
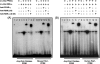



References
-
- Francis, G. A., Fayard, E., Picard, F., and Auwerx, J. (2003) Annu. Rev. Physiol. 65 261-311 - PubMed
-
- Rosen, E. D., and Spiegelman, B. M. (2001) J. Biol. Chem. 276 37731-37734 - PubMed
-
- Picard, F., and Auwerx, J. (2002) Annu. Rev. Nutr. 22 167-197 - PubMed
-
- Lehrke, M., and Lazar, M. A. (2005) Cell 123 992-999 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

